Anders2013: Difference between revisions
| Line 174: | Line 174: | ||
</pre> | </pre> | ||
Note that bowtie2/tophat2 indices for many commonly used reference genomes can be downloaded directly from http://tophat.cbcb.umd.edu/igenomes.html. | Note that bowtie2/tophat2 indices for many commonly used reference genomes can be downloaded directly from http://tophat.cbcb.umd.edu/igenomes.html. | ||
=== Structure of downloaded reference genome from Tophat === | |||
Downloaded genome from tophat takes a long time (20GB for human). Using bowtie2-build program which can takes an enormous time to run. | |||
<pre> | |||
$ tar xzvf Homo_sapiens_Ensembl_GRCh37.tar.gz | |||
$ tree -L 4 Homo_sapiens | |||
Homo_sapiens | |||
└── Ensembl | |||
└── GRCh37 | |||
├── Annotation | |||
│ ├── Archives <==== gtf file | |||
│ ├── Genes -> Archives/archive-current/Genes | |||
│ ├── README.txt -> Archives/archive-current/README.txt | |||
│ ├── SmallRNA -> Archives/archive-current/SmallRNA | |||
│ └── Variation -> Archives/archive-current/Variation | |||
├── GenomeStudio | |||
│ ├── Archives | |||
│ ├── Homo_sapiens -> Archives/archive-2012-03-09-04-49-46/Homo_sapiens | |||
│ └── README.txt | |||
└── Sequence | |||
├── AbundantSequences | |||
├── Bowtie2Index <==== bt2 files | |||
├── BowtieIndex | |||
├── BWAIndex | |||
├── Chromosomes | |||
├── Squashed-Homo_sapiens-Ensembl-GRCh37 | |||
└── WholeGenomeFasta <==== fa file | |||
18 directories, 2 files | |||
$ ls -l Homo_sapiens/Ensembl/GRCh37/Annotation/Archives/archive-2013-03-06-14-23-04/Genes | |||
total 639136 | |||
-rwxrwxr-x 1 brb brb 302 Mar 18 2013 ChromInfo.txt | |||
-rwxrwxr-x 1 brb brb 603096976 Mar 18 2013 genes.gtf | |||
-rwxrwxr-x 1 brb brb 8236175 Mar 18 2013 refFlat.txt.gz | |||
-rwxrwxr-x 1 brb brb 43130092 Mar 18 2013 refGene.txt | |||
</pre> | |||
== Download GTF (gene transfer format) file (77.6 MB) == | == Download GTF (gene transfer format) file (77.6 MB) == | ||
Revision as of 12:05, 2 April 2014
The data is used in the paper "Count-based differential ....." by Anders et al 2013.
Required tools
The version number indicated below is the one I use. It may be updated when you are ready to download that.
- bowtie (bowtie2-2.1.0-linux-x86_64.zip): Mac, Linux, Windows binaries
- samtools (samtools-0.1.19.tar.bz2): equire GCC to compile (build-essential) & zlib1g-dev & libncurses5-dev packages
- tophat (tophat-2.0.10.Linux_x86_64.tar.gz): Linux and Mac binaries
- sra toolkit (sratoolkit.2.3.4-2-ubuntu64.tar.gz): Mac, Ubuntu & CentOS binaries
- HTseq (HTSeq-0.6.1.tar.gz): require Python 2
- IGV (IGV_2.3.26.zip): require Java and registration to download the program
- FastQC (fastqc_v0.10.1.zip): require Java (jdk is needed, jre is not enough)
Installation
If binary executables are available, we don't need to do anything special except adding the path containing the binary to the PATH variable.
If only source code is available, make sure the required tool GCC is installed (Under Ubuntu, we can use sudo apt-get install build-essential). Then we can compile and then install the program by using ./configure, make and make install.
In this exercise, all source code or binary programs were extracted to $HOME/binary/ directory (Use mkdir ~/binary to create this new directory) where each program has its own directory.
The PATH variable
Open ~/.basrhc file using any text editor (such as nano or vi) and add the path containing the binary of each program to PATH variable.
tail ~/.bashrc export PATH=$PATH:~/binary/bowtie2-2.1.0/ export PATH=$PATH:~/binary/tophat-2.0.10.Linux_x86_64/ export PATH=$PATH:~/binary/samtools-0.1.19/ export PATH=$PATH:~/binary/samtools-0.1.19/bcftools/ export PATH=$PATH:~/binary/samtools-0.1.19/misc/ export PATH=$PATH:~/binary/sratoolkit.2.3.4-2-ubuntu64/bin export PATH=$PATH:~/binary/HTSeq-0.6.1/scripts/ export PATH=$PATH:~/binary/IGV_2.3.26/
If we want to make these tools available for ALL users (ie system wide environment), we need to put these lines on /etc/profile file.
Data directory
Download <nprot.2013.099-S1.zip> from the paper's web site and extract it to ~/Anders2013 directory.
brb@brbweb4:~$ mkdir Anders2013 brb@brbweb4:~$ cd Anders2013 brb@brbweb4:~/Anders2013$ wget http://www.nature.com/nprot/journal/v8/n9/extref/nprot.2013.099-S1.zip brb@brbweb4:~/Anders2013$ unzip nprot.2013.099-S1.zip brb@brbweb4:~/Anders2013$ ls CG8144_RNAi-1.count counts.csv README.txt Untreated-1.count Untreated-6.count CG8144_RNAi-3.count __MACOSX samples.csv Untreated-3.count CG8144_RNAi-4.count nprot.2013.099-S1.zip SraRunInfo.csv Untreated-4.count
The raw data is from GSE18508 'modENCODE Drosophila RNA Binding Protein RNAi RNA-Seq Studies'.
Subdirectory
During running tophat program, several subdirectories will be generated. Each subdirectory will contain accepted_hits.bam, junctions.bed, insertions.bed, deletions.bed and other files/sub-subdirectories.
Once samtools program was run, .bam (and .bai) files will be created under Anders2013 directory.
Pipeline
The following summary is extracted from http://www.bioconductor.org/help/course-materials/2013/CSAMA2013/tuesday/afternoon/DESeq_protocol.pdf. It is the same as Anders 2013 paper but the paper requires an access permission.
It would be interesting to monitor the disk space usage before, during and after the execution.
Download example data (22 files in SRA format)
Download Supplement file and read <SraRunInfo.csv> in R. We will choose a subset of data based on "LibraryName" column.
sri = read.csv("SraRunInfo.csv", stringsAsFactors=FALSE)
keep = grep("CG8144|Untreated-",sri$LibraryName)
sri = sri[keep,]
fs = basename(sri$download_path)
for(i in 1:nrow(sri))
download.file(sri$download_path[i], fs[i])
names(sri)
[1] "Run" "ReleaseDate" "spots" "bases"
[5] "avgLength" "size_MB" "AssemblyName" "download_path"
[9] "Experiment" "LibraryName" "LibraryStrategy" "LibrarySelection"
[13] "LibrarySource" "LibraryLayout" "InsertSize" "Platform"
[17] "Model" "SRAStudy" "BioProject" "Study_Pubmed_id"
[21] "ProjectID" "Sample" "BioSample" "TaxID"
[25] "ScientificName" "SampleName" "Submission" "Consent"
apply(sri,2, function(x) length(unique(x)))
Run ReleaseDate spots bases
22 22 22 22
avgLength size_MB AssemblyName download_path
5 22 1 22
Experiment LibraryName LibraryStrategy LibrarySelection
7 7 1 1
LibrarySource LibraryLayout InsertSize Platform
1 2 2 1
Model SRAStudy BioProject Study_Pubmed_id
1 1 1 1
ProjectID Sample BioSample TaxID
1 7 1 1
ScientificName SampleName Submission Consent
1 1 1 1
sri[1:2,]
Run ReleaseDate spots bases avgLength size_MB
1 SRR031714 2010-01-15 10:42:00 5327425 394229450 74 396
2 SRR031715 2010-01-21 14:24:18 5248396 388381304 74 390
AssemblyName
1 NA
2 NA
download_path
1 ftp://ftp-private.ncbi.nlm.nih.gov/sra/sra-instant/reads/ByRun/sra/SRR/SRR031/SRR031714/SRR031714.sra
2 ftp://ftp-private.ncbi.nlm.nih.gov/sra/sra-instant/reads/ByRun/sra/SRR/SRR031/SRR031715/SRR031715.sra
Experiment LibraryName LibraryStrategy LibrarySelection LibrarySource
1 SRX014459 Untreated-3 RNA-Seq cDNA TRANSCRIPTOMIC
2 SRX014459 Untreated-3 RNA-Seq cDNA TRANSCRIPTOMIC
LibraryLayout InsertSize Platform Model SRAStudy
1 PAIRED 200 ILLUMINA Illumina Genome Analyzer II SRP001537
2 PAIRED 200 ILLUMINA Illumina Genome Analyzer II SRP001537
BioProject Study_Pubmed_id ProjectID Sample BioSample
1 GEO Series accession: GSE18508 20921232 168994 SRS008447 NA
2 GEO Series accession: GSE18508 20921232 168994 SRS008447 NA
TaxID ScientificName SampleName Submission Consent
1 7227 Drosophila melanogaster Drosophila melanogaster SRA010243 public
2 7227 Drosophila melanogaster Drosophila melanogaster SRA010243 public
Convert SRA to FASTQ (sra -> fastq)
This takes a long time and it is not run in parallel.
stopifnot( all(file.exists(fs)) ) # assure FTP download was successful
for(f in fs) {
cmd = paste("fastq-dump --split-3", f)
cat(cmd,"\n")
system(cmd) # invoke command
}
fastq-dump --split-3 SRR031714.sra
Read 5327425 spots for SRR031714.sra
Written 5327425 spots for SRR031714.sra
fastq-dump --split-3 SRR031715.sra
Read 5248396 spots for SRR031715.sra
Written 5248396 spots for SRR031715.sra
...
Note that '--split-3' option means legacy 3-file splitting for mate-pairs: First biological reads satisfying dumping conditions are placed in files *_1.fastq and *_2.fastq If only one biological read is present it is placed in *.fastq Biological reads and above are ignored.
brb@brbweb4:~/Anders2013$ ls -lh *.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:47 SRR031708.fastq -rw-r--r-- 1 brb brb 812M Feb 10 12:50 SRR031709.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:56 SRR031710.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 13:02 SRR031711.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:08 SRR031712.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:14 SRR031713.fastq -rw-r--r-- 1 brb brb 1.1G Feb 10 11:48 SRR031714_1.fastq -rw-r--r-- 1 brb brb 1.1G Feb 10 11:48 SRR031714_2.fastq -rw-r--r-- 1 brb brb 1.1G Feb 10 11:54 SRR031715_1.fastq -rw-r--r-- 1 brb brb 1.1G Feb 10 11:54 SRR031715_2.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:02 SRR031716_1.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:02 SRR031716_2.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:10 SRR031717_1.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:10 SRR031717_2.fastq -rw-r--r-- 1 brb brb 1.4G Feb 10 13:21 SRR031718.fastq -rw-r--r-- 1 brb brb 927M Feb 10 13:25 SRR031719.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:32 SRR031720.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:39 SRR031721.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 13:45 SRR031722.fastq -rw-r--r-- 1 brb brb 842M Feb 10 13:49 SRR031723.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:17 SRR031724_1.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:17 SRR031724_2.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:25 SRR031725_1.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:25 SRR031725_2.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:33 SRR031726_1.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:33 SRR031726_2.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:41 SRR031727_1.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:41 SRR031727_2.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:54 SRR031728.fastq -rw-r--r-- 1 brb brb 3.2G Feb 10 14:07 SRR031729.fastq
Download reference genome
wget ftp://ftp.ensembl.org/pub/release-70/fasta/drosophila_melanogaster/dna/Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa.gz gunzip Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa.gz
Note that bowtie2/tophat2 indices for many commonly used reference genomes can be downloaded directly from http://tophat.cbcb.umd.edu/igenomes.html.
Structure of downloaded reference genome from Tophat
Downloaded genome from tophat takes a long time (20GB for human). Using bowtie2-build program which can takes an enormous time to run.
$ tar xzvf Homo_sapiens_Ensembl_GRCh37.tar.gz
$ tree -L 4 Homo_sapiens
Homo_sapiens
└── Ensembl
└── GRCh37
├── Annotation
│ ├── Archives <==== gtf file
│ ├── Genes -> Archives/archive-current/Genes
│ ├── README.txt -> Archives/archive-current/README.txt
│ ├── SmallRNA -> Archives/archive-current/SmallRNA
│ └── Variation -> Archives/archive-current/Variation
├── GenomeStudio
│ ├── Archives
│ ├── Homo_sapiens -> Archives/archive-2012-03-09-04-49-46/Homo_sapiens
│ └── README.txt
└── Sequence
├── AbundantSequences
├── Bowtie2Index <==== bt2 files
├── BowtieIndex
├── BWAIndex
├── Chromosomes
├── Squashed-Homo_sapiens-Ensembl-GRCh37
└── WholeGenomeFasta <==== fa file
18 directories, 2 files
$ ls -l Homo_sapiens/Ensembl/GRCh37/Annotation/Archives/archive-2013-03-06-14-23-04/Genes
total 639136
-rwxrwxr-x 1 brb brb 302 Mar 18 2013 ChromInfo.txt
-rwxrwxr-x 1 brb brb 603096976 Mar 18 2013 genes.gtf
-rwxrwxr-x 1 brb brb 8236175 Mar 18 2013 refFlat.txt.gz
-rwxrwxr-x 1 brb brb 43130092 Mar 18 2013 refGene.txt
Download GTF (gene transfer format) file (77.6 MB)
wget ftp://ftp.ensembl.org/pub/release-70/gtf/drosophila_melanogaster/Drosophila_melanogaster.BDGP5.70.gtf.gz gunzip Drosophila_melanogaster.BDGP5.70.gtf.gz
CRITICAL: Make sure that the gene annotation uses the same coordinate system as the reference FASTA le. Here, both les use BDGP5 (i. e., release 5 of the assembly provided by the Berkeley Drosophila Genome Project), as is apparent from the le names. To be on the safe side here, we recommend to always download the FASTA reference sequence and the GTF annotation data from the same resource provider.
Preprocess reference FASTA files into an index (fasta -> bt2)
Alternatively, pre-built indices for many commonly-used genomes are available from http://tophat.cbcb.umd.edu/igenomes.html <Drosophila_melanogaster_Ensembl_BDGP5.tar.gz> contains genome.1.bt2, genome.2.bt2, genome.3.bt2, genome.fa, genome.rev.1.bt2, and genome.rev.2.bt2 under Drosophila_melanogaster/Ensembl/BDGP5/Sequence/Bowtie2Index/ directory. This takes about 4 minutes 37 seconds on my Core-i7 Laptop computer running xubuntu 12.04.
bowtie2-build -f Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa Dme1_BDGP5_70 brb@brbweb4:~/Anders2013$ ls -lt *.bt2 -rw-r--r-- 1 brb brb 58770281 Feb 10 12:06 Dme1_BDGP5_70.rev.1.bt2 -rw-r--r-- 1 brb brb 40591960 Feb 10 12:06 Dme1_BDGP5_70.rev.2.bt2 -rw-r--r-- 1 brb brb 58770281 Feb 10 12:04 Dme1_BDGP5_70.1.bt2 -rw-r--r-- 1 brb brb 40591960 Feb 10 12:04 Dme1_BDGP5_70.2.bt2 -rw-r--r-- 1 brb brb 339200 Feb 10 12:01 Dme1_BDGP5_70.3.bt2 -rw-r--r-- 1 brb brb 40591953 Feb 10 12:01 Dme1_BDGP5_70.4.bt2
Access quality control through ShortRead or FastQC
library("ShortRead")
fqQC = qa(dirPath=".", pattern=".fastq$", type="fastq")
report(fqQC, type="html", dest="fastqQAreport")
Use a web browser to inspect the generated HTML file (here, stored in the \fastqQAreport" directory) with the quality-assessment report.
Create a metadata table called "samples"
sri$LibraryName = gsub("S2_DRSC_","",sri$LibraryName) # trim label
samples = unique(sri[,c("LibraryName","LibraryLayout")])
for(i in seq_len(nrow(samples))) {
rw = (sri$LibraryName==samples$LibraryName[i])
if(samples$LibraryLayout[i]=="PAIRED") {
samples$fastq1[i] = paste0(sri$Run[rw],"_1.fastq",collapse=",")
samples$fastq2[i] = paste0(sri$Run[rw],"_2.fastq",collapse=",")
} else {
samples$fastq1[i] = paste0(sri$Run[rw],".fastq",collapse=",")
samples$fastq2[i] = ""
}
}
samples$condition = "CTL"
samples$condition[grep("RNAi",samples$LibraryName)] = "KD"
samples$shortname = paste( substr(samples$condition,1,2),
substr(samples$LibraryLayout,1,2), seq_len(nrow(samples)), sep=".")
samples
LibraryName LibraryLayout
1 Untreated-3 PAIRED
3 Untreated-4 PAIRED
5 CG8144_RNAi-3 PAIRED
7 CG8144_RNAi-4 PAIRED
144 Untreated-1 SINGLE
150 CG8144_RNAi-1 SINGLE
156 Untreated-6 SINGLE
fastq1
1 SRR031714_1.fastq,SRR031715_1.fastq
3 SRR031716_1.fastq,SRR031717_1.fastq
5 SRR031724_1.fastq,SRR031725_1.fastq
7 SRR031726_1.fastq,SRR031727_1.fastq
144 SRR031708.fastq,SRR031709.fastq,SRR031710.fastq,SRR031711.fastq,SRR031712.fastq,SRR031713.fastq
150 SRR031718.fastq,SRR031719.fastq,SRR031720.fastq,SRR031721.fastq,SRR031722.fastq,SRR031723.fastq
156 SRR031728.fastq,SRR031729.fastq
fastq2 condition shortname
1 SRR031714_2.fastq,SRR031715_2.fastq CTL CT.PA.1
3 SRR031716_2.fastq,SRR031717_2.fastq CTL CT.PA.2
5 SRR031724_2.fastq,SRR031725_2.fastq KD KD.PA.3
7 SRR031726_2.fastq,SRR031727_2.fastq KD KD.PA.4
144 CTL CT.SI.5
150 KD KD.SI.6
156 CTL CT.SI.7
Align the reads to the reference genome using tophat2 (fastq -> bam, bed)
Note:
- '-p' specifies the number of threads to use, -o specifies the output directory. The first name (bowind) is the name of the index (built in advance).
- It seems even the program cannot find the reference genome, it still runs.
- The run time is about 3 to 4 hours per sample when 7 jobs were running at the same time (5 threads for each sample).
- If only one sample was run it took 45 minutes as the paper said (5 threads). If I specify 1 thread, the running time is 1 hour 44 minutes.
- If I only run 2 jobs using batch, it takes 1 hour and 1 hour 2 mintues to finish these two jobs.
- The input is fastq files. The output is a directory contains 7 files (including *.bam) and 1 subdirectory of logs.
gf = "Drosophila_melanogaster.BDGP5.70.gtf"
bowind = "Dme1_BDGP5_70"
cmd = with(samples, paste("tophat2 -G", gf, "-p 5 -o", LibraryName,
bowind, fastq1, fastq2))
cmd # Actually we want to use system() to run each line of cmd.
[1] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o Untreated-3 Dme1_BDGP5_70 SRR031714_1.fastq,SRR031715_1.fastq SRR031714_2.fastq,SRR031715_2.fastq"
[2] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o Untreated-4 Dme1_BDGP5_70 SRR031716_1.fastq,SRR031717_1.fastq SRR031716_2.fastq,SRR031717_2.fastq"
[3] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o CG8144_RNAi-3 Dme1_BDGP5_70 SRR031724_1.fastq,SRR031725_1.fastq SRR031724_2.fastq,SRR031725_2.fastq"
[4] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o CG8144_RNAi-4 Dme1_BDGP5_70 SRR031726_1.fastq,SRR031727_1.fastq SRR031726_2.fastq,SRR031727_2.fastq"
[5] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o Untreated-1 Dme1_BDGP5_70 SRR031708.fastq,SRR031709.fastq,SRR031710.fastq,SRR031711.fastq,SRR031712.fastq,SRR031713.fastq "
[6] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o CG8144_RNAi-1 Dme1_BDGP5_70 SRR031718.fastq,SRR031719.fastq,SRR031720.fastq,SRR031721.fastq,SRR031722.fastq,SRR031723.fastq "
[7] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o Untreated-6 Dme1_BDGP5_70 SRR031728.fastq,SRR031729.fastq "
Organize, sort and index the BAM files and create SAM files (bam -> sam)
Note
- Potentially we can run the commands in parallel. This step needs lots of disk I/O so it may be better not to run the command in parallel.
- Four steps:
- first step - Input: original aligned bam file. Output: <Untreated-3_sn.bam> (1.5 GB). sort alignment file by read name (-n option).
- Second step - Input: sorted bam file from 1st step. Output: <Untreated-3_sn.sam> (11 GB). BAM<->SAM conversion. The SAM file generated from these 2 steps will be used in HTSEQ-count.
- Third step - Input: original aligned bam file. Output: <Untreated-3_s.bam> (1.2 GB). sort alignment file by chromosomal coordinates.
- Fourth step - Input: sorted bam file from 3rd step. Output: <Untreated-3_s.bam.bai> (375 KB). index alignment. The SAM and BAI files generated from these 2 steps together with GTF file will be used in IGV.
- Examples how to use. See here
for(i in seq_len(nrow(samples))) {
lib = samples$LibraryName[i]
ob = file.path(lib, "accepted_hits.bam")
# sort by name, convert to SAM for htseq-count
cat(paste0("samtools sort -n ",ob," ",lib,"_sn"),"\n")
cat(paste0("samtools view -o ",lib,"_sn.sam ",lib,"_sn.bam"),"\n")
# sort by position and index for IGV
cat(paste0("samtools sort ",ob," ",lib,"_s"),"\n")
cat(paste0("samtools index ",lib,"_s.bam"),"\n\n")
}
samtools sort -n Untreated-3/accepted_hits.bam Untreated-3_sn
samtools view -o Untreated-3_sn.sam Untreated-3_sn.bam
samtools sort Untreated-3/accepted_hits.bam Untreated-3_s
samtools index Untreated-3_s.bam
samtools sort -n Untreated-4/accepted_hits.bam Untreated-4_sn
samtools view -o Untreated-4_sn.sam Untreated-4_sn.bam
samtools sort Untreated-4/accepted_hits.bam Untreated-4_s
samtools index Untreated-4_s.bam
samtools sort -n CG8144_RNAi-3/accepted_hits.bam CG8144_RNAi-3_sn
samtools view -o CG8144_RNAi-3_sn.sam CG8144_RNAi-3_sn.bam
samtools sort CG8144_RNAi-3/accepted_hits.bam CG8144_RNAi-3_s
samtools index CG8144_RNAi-3_s.bam
samtools sort -n CG8144_RNAi-4/accepted_hits.bam CG8144_RNAi-4_sn
samtools view -o CG8144_RNAi-4_sn.sam CG8144_RNAi-4_sn.bam
samtools sort CG8144_RNAi-4/accepted_hits.bam CG8144_RNAi-4_s
samtools index CG8144_RNAi-4_s.bam
samtools sort -n Untreated-1/accepted_hits.bam Untreated-1_sn
samtools view -o Untreated-1_sn.sam Untreated-1_sn.bam
samtools sort Untreated-1/accepted_hits.bam Untreated-1_s
samtools index Untreated-1_s.bam
samtools sort -n CG8144_RNAi-1/accepted_hits.bam CG8144_RNAi-1_sn
samtools view -o CG8144_RNAi-1_sn.sam CG8144_RNAi-1_sn.bam
samtools sort CG8144_RNAi-1/accepted_hits.bam CG8144_RNAi-1_s
samtools index CG8144_RNAi-1_s.bam
samtools sort -n Untreated-6/accepted_hits.bam Untreated-6_sn
samtools view -o Untreated-6_sn.sam Untreated-6_sn.bam
samtools sort Untreated-6/accepted_hits.bam Untreated-6_s
samtools index Untreated-6_s.bam
Inspect alignments with IGV
Count reads using htseq-count (sam -> count)
samples$countf = paste(samples$LibraryName, "count", sep=".")
gf = "Drosophila_melanogaster.BDGP5.70.gtf"
cmd = paste0("htseq-count -s no -a 10 ",
samples$LibraryName, "_sn.sam ",
gf," > ", samples$countf)
cmd
[1] "htseq-count -s no -a 10 Untreated-3_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > Untreated-3.count"
[2] "htseq-count -s no -a 10 Untreated-4_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > Untreated-4.count"
[3] "htseq-count -s no -a 10 CG8144_RNAi-3_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > CG8144_RNAi-3.count"
[4] "htseq-count -s no -a 10 CG8144_RNAi-4_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > CG8144_RNAi-4.count"
[5] "htseq-count -s no -a 10 Untreated-1_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > Untreated-1.count"
[6] "htseq-count -s no -a 10 CG8144_RNAi-1_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > CG8144_RNAi-1.count"
[7] "htseq-count -s no -a 10 Untreated-6_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > Untreated-6.count"
DESeq for DE analysis
> samplesDESeq = with(samples, data.frame(
shortname = I(shortname),
countf = I(countf),
condition = condition,
LibraryLayout = LibraryLayout))
> samplesDESeq
shortname countf condition LibraryLayout
1 CT.PA.1 Untreated-3.count CTL PAIRED
2 CT.PA.2 Untreated-4.count CTL PAIRED
3 KD.PA.3 CG8144_RNAi-3.count KD PAIRED
4 KD.PA.4 CG8144_RNAi-4.count KD PAIRED
5 CT.SI.5 Untreated-1.count CTL SINGLE
6 KD.SI.6 CG8144_RNAi-1.count KD SINGLE
7 CT.SI.7 Untreated-6.count CTL SINGLE
> library("DESeq")
cds <- newCountDataSetFromHTSeqCount(samplesDESeq) # quick, 15682 features, 7 samples
cds <- estimateSizeFactors(cds) # quick
sizeFactors(cds)
# CT.PA.1 CT.PA.2 KD.PA.3 KD.PA.4 CT.SI.5 KD.SI.6 CT.SI.7
# 0.6991612 0.8104921 0.8217403 0.8941097 1.6431467 1.3720872 1.1041186
cds <- estimateDispersions(cds) # quick
res <- nbinomTest(cds,"CTL","KD") # 44 seconds
sum(res$pval <= .05, na.rm=T) # [1] 1574
sum(res$padj <= .05, na.rm=T) # [1] 730
options(width=100)
res[1:3,]
# id baseMean baseMeanA baseMeanB foldChange log2FoldChange pval padj
# 1 FBgn0000003 0.000000 0.000000 0.000000 NaN NaN NA NA
# 2 FBgn0000008 88.526695 88.486694 88.580029 1.001055 0.001520941 0.9298627 1
# 3 FBgn0000014 3.054418 2.327533 4.023599 1.728697 0.789684778 0.6839858 1
Other RNA-Seq data
GEO
Go to http://www.ncbi.nlm.nih.gov/sites/entrez and select 'GEO Datasets' from the drop down menu, and use 'RNA-Seq' as your search term. For example, GSE28666 as used in DAFS paper.
1000 genomes
Human BodyMap 2.0 data from Illumina
- http://www.ensembl.info/blog/2011/05/24/human-bodymap-2-0-data-from-illumina/
- https://usegalaxy.org/u/jeremy/p/galaxy-rna-seq-analysis-exercise. Galaxy needs to install hg19 genome before we can use tophat or bowtie. See Galaxy/Wiki/Admin/Data Preparation page. Genome can be downloaded from tophat webpage. It is 20GB for hg19! We can also download just chromosome 19 fasta file chr19.fa.gz from UCSC genomes FTP server and it is only 19MB. We can then create reference index files (.bt2) by using bowtie2-build command. If we use Galaxy, we don't need to create reference index files.
Other RNA-Seq tools
Galaxy
The first 3 lines will install galaxy on local computer. The last command will start Galaxy. PS. It will take a while to start the first time.
hg clone https://bitbucket.org/galaxy/galaxy-dist/ cd galaxy-dist hg update stable sh run.sh
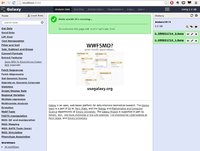
A screenshot of the available tools is given below. Strangely, I did not find "NGS: RNA Analysis" in the NGS: ToolBox on left hand side.
We cannot upload > 2GB fastq data. For such large dataset (we have one 3.3 GB fastq in this case), Galaxy asks to use http or ftp to load the data. I install Apache in Ubuntu and put this fastq file there for upload.
Galaxy does not include bowtie, tophat, .... by default. See this post and Galaxy wiki about the installation. Another option is to modify line 614 of 'universe_wsgl.ini' file and enable ourselves as adminstrator. Also modify line 146 to add a path for installing tools through Galaxy Admin page.
If you submit a lot of jobs (eg each tophat2 generates 5 jobs) some of jobs cannot be run immediately. In my experience, there is a 25 jobs cap. Running jobs has a yellow background color. Waiting for run jobs has a gray background color.
Some helpful information
- https://wiki.galaxyproject.org/Learn Search rna-seq or NGS as keywords.
- Galaxy & DESeq2