T-test
Overview
T-statistic
Let [math]\displaystyle{ \scriptstyle\hat\beta }[/math] be an estimator of parameter β in some statistical model. Then a t-statistic for this parameter is any quantity of the form
- [math]\displaystyle{ t_{\hat{\beta}} = \frac{\hat\beta - \beta_0}{\mathrm{s.e.}(\hat\beta)}, }[/math]
where β0 is a non-random, known constant, and [math]\displaystyle{ \scriptstyle s.e.(\hat\beta) }[/math] is the standard error of the estimator [math]\displaystyle{ \scriptstyle\hat\beta }[/math].
Two sample test assuming equal variance
The t statistic (df = [math]\displaystyle{ n_1 + n_2 - 2 }[/math]) to test whether the means are different can be calculated as follows:
- [math]\displaystyle{ t = \frac{\bar {X}_1 - \bar{X}_2}{s_{X_1X_2} \cdot \sqrt{\frac{1}{n_1}+\frac{1}{n_2}}} }[/math]
where
- [math]\displaystyle{ s_{X_1X_2} = \sqrt{\frac{(n_1-1)s_{X_1}^2+(n_2-1)s_{X_2}^2}{n_1+n_2-2}}. }[/math]
[math]\displaystyle{ s_{X_1X_2} }[/math] is an estimator of the common/pooled standard deviation of the two samples. The square-root of a pooled variance estimator is known as a pooled standard deviation.
- Pooled variance from Wikipedia
- The pooled sample variance is an unbiased estimator of the common variance if Xi and Yi are following the normal distribution.
- (From minitab) The pooled standard deviation is the average spread of all data points about their group mean (not the overall mean). It is a weighted average of each group's standard deviation. The weighting gives larger groups a proportionally greater effect on the overall estimate.
- Type I error rates in two-sample t-test by simulation
Assumptions
The Four Assumptions Made in a T-Test
Two sample test assuming unequal variance
The t statistic (Behrens-Welch test statistic) to test whether the population means are different is calculated as:
- [math]\displaystyle{ t = {\overline{X}_1 - \overline{X}_2 \over s_{\overline{X}_1 - \overline{X}_2}} }[/math]
where
- [math]\displaystyle{ s_{\overline{X}_1 - \overline{X}_2} = \sqrt{{s_1^2 \over n_1} + {s_2^2 \over n_2}}. }[/math]
Here s2 is the unbiased estimator of the variance of the two samples.
The degrees of freedom is evaluated using the Satterthwaite's approximation
- [math]\displaystyle{ df = { ({s_1^2 \over n_1} + {s_2^2 \over n_2})^2 \over {({s_1^2 \over n_1})^2 \over n_1-1} + {({s_2^2 \over n_2})^2 \over n_2-1} }. }[/math]
See an example The Satterthwaite Approximation: Definition & Example.
Paired test, Wilcoxon signed rank test
- Have you ever asked yourself, "how should I approach the classic pre-post analysis?"
- How to best visualize one-sample test?
- Can R visualize the t.test or other hypothesis test results?, gginference package
- Wilcoxon Test in R
- How Do We Perform a Paired t-Test When We Don’t Know How to Pair? 2022
Z-value/Z-score
If the population parameters are known, then rather than computing the t-statistic, one can compute the z-score.
Nonparametric test: Wilcoxon rank sum test
Mann–Whitney U test/Wilcoxon rank-sum test or Wilcoxon–Mann–Whitney test
Sensitive to differences in location
Wilcoxon test in R: how to compare 2 groups under the non-normality assumption using wilcox.test()
Wilcoxon-Mann-Whitney test and a small sample size. if performing a WMW test comparing S1=(1,2) and S2=(100,300) it wouldn’t differ of comparing S1=(1,2) and S2=(4,5). Therefore when having a small sample size this is a great loss of information.
Nonparametric test: Kolmogorov-Smirnov test
Sensitive to difference in shape and location of the distribution functions of two groups
Kolmogorov–Smirnov test. The Kolmogorov–Smirnov statistic for a given cumulative distribution function F(x) is
- [math]\displaystyle{ D_n= \sup_x |F_n(x)-F(x)| }[/math]
where supx is the supremum of the set of distances.
Gene set enrichment analysis The Enrichment score ES is is the maximum deviation from zero of P(hit)-P(miss). For a randomly distributed S, ES will be relatively small, but if it is concentrated at the top or bottom of the list, or otherwise nonrandomly distributed, then ES(S) will be correspondingly high. When p=0, ES reduces to the standard Kolmogorov–Smirnov statistic. c; when p=1, we are weighting the genes in S by their correlation with C normalized by the sum of the correlations over all of the genes in S.
- [math]\displaystyle{ P_{hit}(S, i) = \sum \frac{|r_j|^p}{N_R}, P_{miss}(S, i)= \sum \frac{1}{(N-N_H)} }[/math] where [math]\displaystyle{ N_R = \sum_{g_j \in S} |r_j|^p }[/math].
See also Statistics -> GSEA.
Limma: Empirical Bayes method
- Some Bioconductor packages: limma, RnBeads, IMA, minfi packages.
- The moderated T-statistics used in Limma is defined on Limma's user guide.
- Diagram of usage ?makeContrasts, ?contrasts.fit, ?eBayes
lmFit contrasts.fit eBayes topTable x ------> fit ------------------> fit2 -----> fit2 ---------> ^ ^ | | model.matrix | makeContrasts | class ---------> design ----------> contrasts - Moderated t-test mod.t.test() from MKmisc package
- Examples of contrasts (search contrasts.fit and/or model.matrix from the user guide)
# Ex 1 (Single channel design): design <- model.matrix(~ 0+factor(c(1,1,1,2,2,3,3,3))) # number of samples x number of groups colnames(design) <- c("group1", "group2", "group3") fit <- lmFit(eset, design) contrast.matrix <- makeContrasts(group2-group1, group3-group2, group3-group1, levels=design) # number of groups x number of contrasts fit2 <- contrasts.fit(fit, contrast.matrix) fit2 <- eBayes(fit2) topTable(fit2, coef=1, adjust="BH") topTable(fit2, coef=1, sort = "none", n = Inf, adjust="BH")$adj.P.Val # Ex 2 (Common reference design): targets <- readTargets("runxtargets.txt") design <- modelMatrix(targets, ref="EGFP") contrast.matrix <- makeContrasts(AML1,CBFb,AML1.CBFb,AML1.CBFb-AML1,AML1.CBFb-CBFb, levels=design) fit <- lmFit(MA, design) fit2 <- contrasts.fit(fit, contrasts.matrix) fit2 <- eBayes(fit2) # Ex 3 (Direct two-color design): design <- modelMatrix(targets, ref="CD4") contrast.matrix <- cbind("CD8-CD4"=c(1,0),"DN-CD4"=c(0,1),"CD8-DN"=c(1,-1)) rownames(contrast.matrix) <- colnames(design) fit <- lmFit(eset, design) fit2 <- contrasts.fit(fit, contrast.matrix) # Ex 4 (Single channel + Two groups): fit <- lmFit(eset, design) cont.matrix <- makeContrasts(MUvsWT=MU-WT, levels=design) fit2 <- contrasts.fit(fit, cont.matrix) fit2 <- eBayes(fit2) # Ex 5 (Single channel + Several groups): f <- factor(targets$Target, levels=c("RNA1","RNA2","RNA3")) design <- model.matrix(~0+f) colnames(design) <- c("RNA1","RNA2","RNA3") fit <- lmFit(eset, design) contrast.matrix <- makeContrasts(RNA2-RNA1, RNA3-RNA2, RNA3-RNA1, levels=design) fit2 <- contrasts.fit(fit, contrast.matrix) fit2 <- eBayes(fit2) # Ex 6 (Single channel + Interaction models 2x2 Factorial Designs) : cont.matrix <- makeContrasts( SvsUinWT=WT.S-WT.U, SvsUinMu=Mu.S-Mu.U, Diff=(Mu.S-Mu.U)-(WT.S-WT.U), levels=design) fit2 <- contrasts.fit(fit, cont.matrix) fit2 <- eBayes(fit2) - Example from user guide 17.3 (Mammary progenitor cell populations)
setwd("~/Downloads/IlluminaCaseStudy") url <- c("http://bioinf.wehi.edu.au/marray/IlluminaCaseStudy/probe%20profile.txt.gz", "http://bioinf.wehi.edu.au/marray/IlluminaCaseStudy/control%20probe%20profile.txt.gz", "http://bioinf.wehi.edu.au/marray/IlluminaCaseStudy/Targets.txt") for(i in url) system(paste("wget ", i)) system("gunzip probe%20profile.txt.gz") system("gunzip control%20probe%20profile.txt.gz") source("http://www.bioconductor.org/biocLite.R") biocLite("limma") biocLite("statmod") library(limma) targets <- readTargets() targets x <- read.ilmn(files="probe profile.txt",ctrlfiles="control probe profile.txt", other.columns="Detection") options(digits=3) head(x$E) boxplot(log2(x$E),range=0,ylab="log2 intensity") y <- neqc(x) dim(y) expressed <- rowSums(y$other$Detection < 0.05) >= 3 y <- y[expressed,] dim(y) # 24691 12 plotMDS(y,labels=targets$CellType) ct <- factor(targets$CellType) design <- model.matrix(~0+ct) colnames(design) <- levels(ct) dupcor <- duplicateCorrelation(y,design,block=targets$Donor) # need statmod dupcor$consensus.correlation fit <- lmFit(y, design, block=targets$Donor, correlation=dupcor$consensus.correlation) contrasts <- makeContrasts(ML-MS, LP-MS, ML-LP, levels=design) fit2 <- contrasts.fit(fit, contrasts) fit2 <- eBayes(fit2, trend=TRUE) summary(decideTests(fit2, method="global")) topTable(fit2, coef=1) # Top ten differentially expressed probes between ML and MS # SYMBOL TargetID logFC AveExpr t P.Value adj.P.Val B # ILMN_1766707 IL17B <NA> -4.19 5.94 -29.0 2.51e-12 5.19e-08 18.1 # ILMN_1706051 PLD5 <NA> -4.00 5.67 -27.8 4.20e-12 5.19e-08 17.7 # ... tT <- topTable(fit2, coef=1, number = Inf) dim(tT) # [1] 24691 8 - Three groups comparison (What is the difference of A vs Other AND A vs (B+C)/2?). Contrasts comparing one factor to multiple others
library(limma) set.seed(1234) n <- 100 testexpr <- matrix(rnorm(n * 10, 5, 1), nc= 10) testexpr[, 6:7] <- testexpr[, 6:7] + 7 # mean is 12 design1 <- model.matrix(~ 0 + as.factor(c(rep(1,5),2,2,3,3,3))) design2 <- matrix(c(rep(1,5),rep(0,5),rep(0,5),rep(1,5)),ncol=2) colnames(design1) <- LETTERS[1:3] colnames(design2) <- c("A", "Other") fit1 <- lmFit(testexpr,design1) contrasts.matrix1 <- makeContrasts("AvsOther"=A-(B+C)/2, levels = design1) fit1 <- eBayes(contrasts.fit(fit1,contrasts=contrasts.matrix1)) fit2 <- lmFit(testexpr,design2) contrasts.matrix2 <- makeContrasts("AvsOther"=A-Other, levels = design2) fit2 <- eBayes(contrasts.fit(fit2,contrasts=contrasts.matrix2)) t1 <- topTable(fit1,coef=1, number = Inf) t2 <- topTable(fit2,coef=1, number = Inf) rbind(head(t1, 3), tail(t1, 3)) # logFC AveExpr t P.Value adj.P.Val B # 92 -5.293932 5.810926 -8.200138 1.147084e-15 1.147084e-13 26.335702 # 81 -5.045682 5.949507 -7.815607 2.009706e-14 1.004853e-12 23.334600 # 37 -4.720906 6.182821 -7.312539 7.186627e-13 2.395542e-11 19.625964 # 27 -2.127055 6.854324 -3.294744 1.034742e-03 1.055859e-03 -1.141991 # 86 -1.938148 7.153142 -3.002133 2.776390e-03 2.804434e-03 -2.039869 # 75 -1.876490 6.516004 -2.906626 3.768951e-03 3.768951e-03 -2.314869 rbind(head(t2, 3), tail(t2, 3)) # logFC AveExpr t P.Value adj.P.Val B # 92 -4.518551 5.810926 -2.5022436 0.01253944 0.2367295 -4.587080 # 81 -4.500503 5.949507 -2.4922492 0.01289503 0.2367295 -4.587156 # 37 -4.111158 6.182821 -2.2766414 0.02307100 0.2367295 -4.588728 # 27 -1.496546 6.854324 -0.8287440 0.40749644 0.4158127 -4.595601 # 86 -1.341607 7.153142 -0.7429435 0.45773401 0.4623576 -4.595807 # 75 -1.171366 6.516004 -0.6486690 0.51673851 0.5167385 -4.596008 var(as.numeric(testexpr[, 6:10])) # [1] 12.38074 var(as.numeric(testexpr[, 6:7])) # [1] 0.8501378 var(as.numeric(testexpr[, 8:10])) # [1] 0.9640699As we can see the p-values returned from the first contrast are very small (large mean but small variance) but the p-values returned from the 2nd contrast are large (still large mean but very large variance). The variance from the "Other" group can be calculated from a mixture distribution ( pdf = .4 N(12, 1) + .6 N(5, 1), VarY = E(Y^2) - (EY)^2 where E(Y^2) = .4 (VarX1 + (EX1)^2) + .6 (VarX2 + (EX2)^2) = 73.6 and EY = .4 * 12 + .6 * 5 = 7.8; so VarY = 73.6 - 7.8^2 = 12.76). - Correct assumptions of using limma moderated t-test and the paper Should We Abandon the t-Test in the Analysis of Gene Expression Microarray Data: A Comparison of Variance Modeling Strategies.
- Evaluation: statistical power (figure 3, 4, 5), false-positive rate (table 2), execution time and ease of use (table 3)
- Limma presents several advantages
- RVM inflates the expected number of false-positives when sample size is small. On the other hand the, RVM is very close to Limma from either their formulas (p3 of the supporting info) or the Hierarchical clustering (figure 2) of two examples.
- Slides
- Use Limma to run ordinary T tests, Limma Moderated and Ordinary t-statistics
# where 'fit' is the output from lmFit() or contrasts.fit(). unmod.t <- fit$coefficients/fit$stdev.unscaled/fit$sigma pval <- 2*pt(-abs(unmod.t), fit$df.residual) # Following the above example t.test(testexpr[1, 1:5], testexpr[1, 6:10], var.equal = T) # Two Sample t-test # # data: testexpr[1, 1:5] and testexpr[1, 6:10] # t = -1.2404, df = 8, p-value = 0.25 # alternative hypothesis: true difference in means is not equal to 0 # 95 percent confidence interval: # -7.987791 2.400082 # sample estimates: # mean of x mean of y # 4.577183 7.371037 fit2$coefficients[1] / (fit2$stdev.unscaled[1] * fit2$sigma[1]) # Ordinary t-statistic # [1] -1.240416 fit2$coefficients[1] / (fit2$stdev.unscaled[1] * sqrt(fit2$s2.post[1])) # moderated t-statistic # [1] -1.547156 topTable(fit2,coef=1, sort.by = "none")[1,] # logFC AveExpr t P.Value adj.P.Val B # 1 -2.793855 5.974110 -1.547156 0.1222210 0.2367295 -4.592992 # Square root of the pooled variance fit2$sigma[1] # [1] 3.561284 (((5-1)*var(testexpr[1, 1:5]) + (5-1)*var(testexpr[1, 6:10]))/(5+5-2)) %>% sqrt() # [1] 3.561284
- Comparison of ordinary T-statistic, RVM T-statistic and Limma/eBayes moderated T-statistic.
| Test statistic for gene g | ||
|---|---|---|
| Ordinary T-test | [math]\displaystyle{ \frac{\overline{y}_{g1} - \overline{y}_{g2}}{S_g^{Pooled}/\sqrt{1/n_1 + 1/n_2}} }[/math] | [math]\displaystyle{ (S_g^{Pooled})^2 = \frac{(n_1-1)S_{g1}^2 + (n_2-1)S_{g2}^2}{n1+n2-2} }[/math] |
| RVM | [math]\displaystyle{ \frac{\overline{y}_{g1} - \overline{y}_{g2}}{S_g^{RVM}/\sqrt{1/n_1 + 1/n_2}} }[/math] | [math]\displaystyle{ (S_g^{RVM})^2 = \frac{(n_1+n_2-2)S_{g}^2 + 2*a*(a*b)^{-1}}{n1+n2-2+2*a} }[/math] |
| Limma | [math]\displaystyle{ \frac{\overline{y}_{g1} - \overline{y}_{g2}}{S_g^{Limma}/\sqrt{1/n_1 + 1/n_2}} }[/math] | [math]\displaystyle{ (S_g^{Limma})^2 = \frac{d_0 S_0^2 + d_g S_g^2}{d_0 + d_g} }[/math] |
- In Limma,
- [math]\displaystyle{ \sigma_g^2 }[/math] assumes an inverse Chi-square distribution with mean [math]\displaystyle{ S_0^2 }[/math] and [math]\displaystyle{ d_0 }[/math] degrees of freedom
- [math]\displaystyle{ d_0 }[/math] (fit$df.prior) and [math]\displaystyle{ d_g }[/math] are, respectively, prior and residual/empirical degrees of freedom.
- [math]\displaystyle{ S_0^2 }[/math] (fit$s2.prior) is the prior distribution and [math]\displaystyle{ S_g^2 }[/math] is the pooled variance.
- [math]\displaystyle{ (S_g^{Limma})^2 }[/math] can be obtained from fit$s2.post.
- Empirical Bayes estimation of normal means, accounting for uncertainty in estimated standard errors Lu 2019
t.test() vs aov() vs sva::f.pvalue() vs genefilter::rowttests()
- t.test() & aov() & sva::f.pvalue() & AT function will be equivalent if we assume equal variances in groups (not the default in t.test)
- See examples in gist.
# Method 1:
tmp <- data.frame(groups=groups.gem,
x=combat_edata3["TDRD7", 29:40])
anova(aov(x ~ groups, data = tmp))
# Analysis of Variance Table
#
# Response: x
# Df Sum Sq Mean Sq F value Pr(>F)
# groups 1 0.0659 0.06591 0.1522 0.7047
# Method 2:
t.test(combat_edata3["TDRD7", 29:40][groups.gem == "CR"],
combat_edata3["TDRD7", 29:40][groups.gem == "PD"])
# 0.7134
t.test(combat_edata3["TDRD7", 29:40][groups.gem == "CR"],
combat_edata3["TDRD7", 29:40][groups.gem == "PD"], var.equal = TRUE)
# 0.7047
# Method 3:
require(sva)
pheno <- data.frame(groups.gem = groups.gem)
mod = model.matrix(~as.factor(groups.gem), data = pheno)
mod0 = model.matrix(~1, data = pheno)
f.pvalue(combat_edata3[c("TDRD7", "COX7A2"), 29:40],
mod, mod0)
# TDRD7 COX7A2
# 0.7046624 0.2516682
# Method 4:
# load some functions from AT randomForest plugin
tmp2 <- Vat2(combat_edata3[c("TDRD7", "COX7A2"), 29:40],
ifelse(groups.gem == "CR", 0, 1),
grp=c(0,1), rvm = FALSE)
tmp2$tp
# TDRD7 COX7A2
# 0.7046624 0.2516682
# Method 5: https://stats.stackexchange.com/a/474335
# library(genefilter)
# Method 6:
# library(limma)
sapply + lm()
using lm() in R for a series of independent fits. Gene expression is on response part (Y). We have nG columns on Y. We'll fit a regression for each column of Y and the same X.
Treatment effect estimation
Permutation test
- When Your Permutation Test is Doomed to Fail
- Permutation P-values should never be zero: calculating exact P-values when permutations are randomly drawn Phipson & Smyth 2010
- getPvals() from singscore package. The null hypothesis is that the gene-set is not enriched in the sample. Question: it seems the permutation p-value is wrong. It should be calculated by using the tail values not by one-sided because the scores can be positive or negative.
# observed: 1 x nsamples # permuteResults: B x nsamples # pvals: 1 x nsamples pvals <- colSums(permuteResult > matrix(1, nrow = nrow(permuteResult), ncol = 1) %*% observed) / B # Consider B=1000, nsamples=5 # e.g. pvals = (0.996, 0.000, 0.999, 0.000, 0.530) pvals <- sapply(pvals, max, 1/B) # e.g. pvals = (0.996, 0.001, 0.999, 0.001, 0.530) range(permuteResult[,1]) # [1] -0.09813482 0.10853650 round(scoredf[,"TotalScore"], 3) # [1] -0.088 0.287 -0.099 0.271 -0.002 0.176 0.017 0.188 -0.062 -0.065 - In ArrayTools, LS score (= -sum (log p_j)) is always > 0. So the permutation p-value is calculated using one-sided.
- GSA package. Correct p-value in GSA (Gene set enrichment) permutation tests? Why it uses one-sided?
- Two-sided permutation test vs. two one-sided
- https://faculty.washington.edu/kenrice/sisg/SISG-08-06.pdf
- How about the fgsea package?
- Roughly estimate the number of permutations based on the desired FDR GSEA for RNA-seq analysis
ANOVA
- Practical Regression and Anova using R by Julian J. Faraway, 2002
- A simple ANOVA
- Repeated measures ANOVA in R Exercises
- Mixed models for ANOVA designs with one observation per unit of observation and cell of the design
- afex package, afex_plot(): Publication-Ready Plots for Factorial Designs
- Experiment designs for Agriculture
- ANOVA in R from statsandr.com
- Design and Analysis of Experiments by Montgomery
- https://cran.r-project.org/web/views/ExperimentalDesign.html
Partition of sum of squares
https://en.wikipedia.org/wiki/Partition_of_sums_of_squares
F-test in anova
How is the F-statistic computed in anova() when there are multiple models?
Common tests are linear models
https://lindeloev.github.io/tests-as-linear/
ANOVA Vs Multiple Comparisons
Post-hoc test
Determine which levels have significantly different means.
- http://jamesmarquezportfolio.com/one_way_anova_with_post_hocs_in_r.html
- pairwise.t.test() for one-way ANOVA
- Post-hoc Pairwise Comparisons of Two-way ANOVA using TukeyHSD().
- post-hoc tests: pairwise.t.test versus TukeyHSD test
- How to Perform Dunnett’s Test in R
- How to do Pairwise Comparisons in R?
- [https://www.statology.org/tukey-vs-bonferroni-vs-scheffe/
TukeyHSD (Honestly Significant Difference), diagnostic checking
https://datascienceplus.com/one-way-anova-in-r/, Tukey HSD for Post-Hoc Analysis (detailed explanation including the type 1 error problem in multiple testings)
- TukeyHSD for the pairwise tests
- You can’t just perform a series of t tests, because that would greatly increase your likelihood of a Type I error.
- compute something analogous to a t score for each pair of means, but you don’t compare it to the Student’s t distribution. Instead, you use a new distribution called the studentized range (from Wikipedia) or q distribution.
- Suppose that we take a sample of size n from each of k populations with the same normal distribution N(μ, σ) and suppose that [math]\displaystyle{ \bar{y} }[/math]min is the smallest of these sample means and [math]\displaystyle{ \bar{y} }[/math]max is the largest of these sample means, and suppose S2 is the pooled sample variance from these samples. Then the following random variable has a Studentized range distribution: [math]\displaystyle{ q = \frac{\overline{y}_{\max} - \overline{y}_{\min}}{S/\sqrt{n}} }[/math]
- One-Way ANOVA Test in R from sthda.com.
res.aov <- aov(weight ~ group, data = PlantGrowth) summary(res.aov) # Df Sum Sq Mean Sq F value Pr(>F) # group 2 3.766 1.8832 4.846 0.0159 * # Residuals 27 10.492 0.3886 TukeyHSD(res.aov) # Tukey multiple comparisons of means # 95% family-wise confidence level # # Fit: aov(formula = weight ~ group, data = PlantGrowth) # # $group # diff lwr upr p adj # trt1-ctrl -0.371 -1.0622161 0.3202161 0.3908711 # trt2-ctrl 0.494 -0.1972161 1.1852161 0.1979960 # trt2-trt1 0.865 0.1737839 1.5562161 0.0120064 # Extra: # Check your data my_data <- PlantGrowth levels(my_data$group) set.seed(1234) dplyr::sample_n(my_data, 10) # compute the summary statistics by group library(dplyr) group_by(my_data, group) %>% summarise( count = n(), mean = mean(weight, na.rm = TRUE), sd = sd(weight, na.rm = TRUE) ) - Or we can use Benjamini-Hochberg method for p-value adjustment in pairwise comparisons
library(multcomp) pairwise.t.test(my_data$weight, my_data$group, p.adjust.method = "BH") # ctrl trt1 # trt1 0.194 - # trt2 0.132 0.013 # # P value adjustment method: BH
- Shapiro-Wilk test for normality
# Extract the residuals aov_residuals <- residuals(object = res.aov ) # Run Shapiro-Wilk test shapiro.test(x = aov_residuals )
- Bartlett test and Levene test for the homogeneity of variances across the groups
Repeated measure
- How to do Repeated Measures ANOVAs in R
- Cross-over Repeated Measure Designs
- Cross-over study design with a major constraint
- Repeated Measures of ANOVA in R Complete Tutorial
Combining insignificant factor levels
COMBINING AUTOMATICALLY FACTOR LEVELS IN R
Omnibus tests
- https://en.wikipedia.org/wiki/Omnibus_test
- Understanding the definition of omnibus tests Tests are refereed to as omnibus if after rejecting the null hypothesis you do not know where the differences assessed by the statistical test are. In the case of F tests they are omnibus when there is more than one df in the numerator (3 or more groups) it is omnibus.
One-way ANOVA
https://www.mathstat.dal.ca/~stat2080/Fall14/Lecturenotes/anova1.pdf
Two-way ANOVA
- Interpreting Interactions when Main Effects are Not Significant
- Actually, you can interpret some main effects in the presence of an interaction
- Confusion about Deseq2 wording in the vignette (additive model with main effects only vs interaction)
- Why and When to Include Interactions in a Regression Model
- they have large main effects
- the effect of one changes for various subgroups of the other
- the interaction has been proven in previous studies
- you want to explore new hypotheses
- How to interpret main effects when the interaction effect is not significant?
- When Main Effects are Not Significant, But the Interaction Is. A cross-over interaction.
- Main effects are not significant anymore after adding interaction terms in my linear regression. Frank Harrell answered.
- Can the interaction term of two insignificant coefficients be significant?
- Why does the main effect become not significant when I add an interaction term in a regression model? The interaction effect is more influential in explaining differences in the outcome variable than either the "main effect". moderator
- Including an Interaction term leads to insignificant direct effects. The "main effects" no longer carry the same meaning when an interaction term is included in the model, so their coefficients have no necessary relationship to the coefficients seen by the variables of the same name in a no-interaction model.
- How to Create an Interaction Plot in R? stats::interaction.plot()
An example
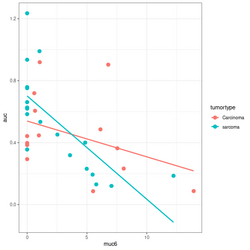
devtools::source_gist("https://gist.github.com/arraytools/88449e3c92c752eb7a66ee0189b09606")
dim(df) # [1] 33 3
df[1:2, ]
# muc6 tumortype auc
# V1 0.6599192 Carcinoma 0.6056
# V2 12.2342844 sarcoma 0.1858
summary(lm(auc~muc6, data=df))
# Coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 0.63065 0.05388 11.706 6.56e-13 ***
# muc6 -0.04367 0.01119 -3.903 0.000478 ***
# Residual standard error: 0.2407 on 31 degrees of freedom
# Multiple R-squared: 0.3295, Adjusted R-squared: 0.3079
# F-statistic: 15.23 on 1 and 31 DF, p-value: 0.000478
summary(lm(auc~., data=df))
# Coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 0.61262 0.07764 7.891 8.32e-09 ***
# muc6 -0.04312 0.01148 -3.758 0.000739 ***
# tumortypesarcoma 0.02846 0.08697 0.327 0.745791
# Residual standard error: 0.2443 on 30 degrees of freedom
# Multiple R-squared: 0.3319, Adjusted R-squared: 0.2873
# F-statistic: 7.451 on 2 and 30 DF, p-value: 0.00236
summary(lm(auc~muc6*tumortype, data=df))
# Coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 0.53938 0.08281 6.514 3.93e-07 ***
# muc6 -0.02312 0.01490 -1.552 0.1316
# tumortypesarcoma 0.16194 0.10690 1.515 0.1406
# muc6:tumortypesarcoma -0.04356 0.02198 -1.982 0.0571 .
# Residual standard error: 0.2332 on 29 degrees of freedom
# Multiple R-squared: 0.4116, Adjusted R-squared: 0.3507
# F-statistic: 6.761 on 3 and 29 DF, p-value: 0.001352
library(ggplot2); library(magrittr)
df %>% ggplot(aes(muc6, auc, col=tumortype)) +
geom_point(size=3) +
geom_smooth(method="lm",se = FALSE)
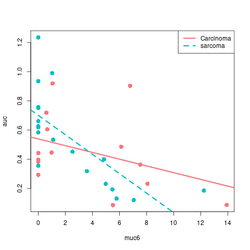
# Base R plot version
col2 <- c("#F8767D", "#00BFC4") # see my ggplot2 page
col <- col2[as.integer(factor(df$tumortype))]
f$tumortype[1:5]
# [1] "Carcinoma" "sarcoma" "Carcinoma" "Carcinoma" "sarcoma"
as.integer(factor(df$tumortype))[1:5]
# [1] 1 2 1 1 2
plot(df$muc6, df$auc, col = col, xlab="muc6", ylab="auc", pch=16, cex=1.5)
abline(a = coefs[1], b=coefs[2], col = col2[1], lwd=3)
abline(a = coefs[1] + coefs[3], b = coefs[2] + coefs[4], lty=2, col = col[2], lwd=3)
legend("topright", c("Carcinoma", "sarcoma"), col = col2, lty=1:2, lwd=3)
Type I, II and III Sums of Squares
- Type I, II and III Sums of Squares – the explanation
- type I (sequential): SS(A) for factor A. SS(B | A) for factor B. SS(AB | B, A) for interaction AB. If the data is unbalanced, you will get a different result of using anova(lm(y ~ A * B, data=search)) & anova(lm(y ~ B * A, data=search)). This is default in R.
- type II (ADDED AFTER OTHER MAIN EFFECTS): SS(A | B) for factor A. SS(B | A) for factor B. R's anova().
- type III (ADDED LAST): SS(A | B, AB) for factor A. SS(B | A, AB) for factor B. This is the default in SAS. In R, car:::Anova(, type="III").
- Everything You Always Wanted to Know About ANOVA
- It seems the order does not make a difference in linear regression or Cox regression.
Kruskal-Wallis one-way analysis of variance
- https://en.wikipedia.org/wiki/Kruskal%E2%80%93Wallis_one-way_analysis_of_variance
- How to perform the Kruskal-Wallis test in R?
- Kruskal Wallis test in R-One-way ANOVA Alternative
Multilevel, Simpson paradox
Multilevel Correlations: A New Method for Common Problems. Simpson paradox.
ANCOVA
MANOVA
- MANOVA(Multivariate Analysis of Variance) using R
- MANOVA in R – How To Implement and Interpret One-Way MANOVA