Venn diagram: Difference between revisions
Appearance
Created page with "* [https://cran.r-project.org/web/packages/eulerr/index.html eulerr] package * [https://codingenes.wordpress.com/2013/09/30/visualization-biological-data-using-proportional-ve..." |
|||
| (7 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
* [https:// | = Misc = | ||
* 2014 Paper [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0101717 eulerAPE: Drawing Area-Proportional 3-Venn Diagrams Using Ellipses] which includes an overview of several tools. The paper is linked from the website https://upset.app. | |||
* [https://codingenes.wordpress.com/2013/09/30/visualization-biological-data-using-proportional-venn-diagrams/ Visualization biological data using proportional venn diagrams] | * [https://codingenes.wordpress.com/2013/09/30/visualization-biological-data-using-proportional-venn-diagrams/ Visualization biological data using proportional venn diagrams] | ||
* [https://www.datanovia.com/en/blog/venn-diagram-with-r-or-rstudio-a-million-ways/#using-the-venndiagram-r-package VENN DIAGRAM WITH R OR RSTUDIO: A MILLION WAYS] | * [https://www.datanovia.com/en/blog/venn-diagram-with-r-or-rstudio-a-million-ways/#using-the-venndiagram-r-package VENN DIAGRAM WITH R OR RSTUDIO: A MILLION WAYS] | ||
<ul> | <ul> | ||
<li> | <li>http://manuals.bioinformatics.ucr.edu/home/R_BioCondManual#TOC-Venn-Diagrams and the [http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/overLapper.R R code] or the [http://www.bioconductor.org/packages/release/bioc/html/systemPipeR.html Bioc package systemPipeR] | ||
{{Pre}} | |||
# systemPipeR package method | |||
library(systemPipeR) | |||
setlist <- list(A=sample(letters, 18), B=sample(letters, 16), C=sample(letters, 20), D=sample(letters, 22), E=sample(letters, 18)) | |||
OLlist <- overLapper(setlist[1:3], type="vennsets") | |||
vennPlot(list(OLlist)) | |||
# R script source method | |||
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/overLapper.R") | |||
setlist <- list(A=sample(letters, 18), B=sample(letters, 16), C=sample(letters, 20), D=sample(letters, 22), E=sample(letters, 18)) | |||
# or (obtained by dput(setlist)) | |||
setlist <- structure(list(A = c("o", "h", "u", "p", "i", "s", "a", "w", | |||
"b", "z", "n", "c", "k", "j", "y", "m", "t", "q"), B = c("h", | |||
"r", "x", "y", "b", "t", "d", "o", "m", "q", "g", "v", "c", "u", | |||
"f", "z"), C = c("b", "e", "t", "u", "s", "j", "o", "k", "d", | |||
"l", "g", "i", "w", "n", "p", "a", "y", "x", "m", "z"), D = c("f", | |||
"g", "b", "k", "j", "m", "e", "q", "i", "d", "o", "l", "c", "t", | |||
"x", "r", "s", "u", "w", "a", "z", "n"), E = c("u", "w", "o", | |||
"k", "n", "h", "p", "z", "l", "m", "r", "d", "q", "s", "x", "b", | |||
"v", "t"), F = c("o", "j", "r", "c", "l", "l", "u", "b", "f", | |||
"d", "u", "m", "y", "t", "y", "s", "a", "g", "t", "m", "x", "m" | |||
)), .Names = c("A", "B", "C", "D", "E", "F")) | |||
OLlist <- overLapper(setlist[1:3], type="vennsets") | |||
counts <- list(sapply(OLlist$Venn_List, length)) | |||
vennPlot(counts=counts) | |||
</pre> | |||
[[:File:Vennplot.png]] | |||
</li> | |||
</ul> | |||
== Online tools == | |||
* https://molbiotools.com/listcompare.php | |||
= UpSet plot = | |||
* https://upset.app/ | |||
* [https://cran.r-project.org/web/packages/UpSetR/index.html UpSetR] | |||
* [https://cran.r-project.org/web/packages/ComplexUpset/index.html ComplexUpset]: Create Complex UpSet Plots Using 'ggplot2' Components | |||
* [https://jokergoo.github.io/ComplexHeatmap-reference/book/upset-plot.html Chapter 8 UpSet plot] | |||
= venn = | |||
* [https://cran.r-project.org/web/packages/venn/index.html venn] package (best so far) | |||
** [https://github.com/dusadrian/venn/blob/master/R/plotRules.R#L209 polygon()] function call. | |||
* [https://statisticsglobe.com/venn-r-package Introduction to the venn Package in R (6 Examples) | How to Draw Up to 7 Sets] | |||
<pre> | <pre> | ||
library(venn) | |||
set.seed(12345) | |||
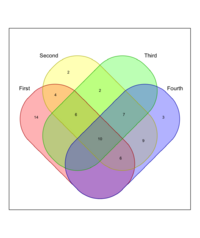
x <- list(First = 1:40, Second = 15:60, Third = sample(25:50, 25), | |||
Fourth=sample(15:65, 35)) | |||
venn(x, ilabels = "counts", zcolor = "style", ilcs = 1.2, sncs = 1) | |||
</pre> | </pre> | ||
In this example, '''ilcs''' and '''sncs''' will control the font size of the '''intersection labels''' and '''set names''', respectively. | |||
[[File:Venn4.png|200px]] | |||
== ggVennDiagram == | |||
https://cloud.r-project.org/web/packages/ggVennDiagram/index.html, [https://gaospecial.github.io/ggVennDiagram/ github]. | |||
Cons: there is no way to have multiple colors for different sets. | |||
= VennDiagram = | |||
<ul> | <ul> | ||
<li>[https://cran.r-project.org/web/packages/VennDiagram/ VennDiagram]. As good as the "venn" package. Many reverse imports and suggests. | |||
<li>[https://www.datanovia.com/en/blog/venn-diagram-with-r-or-rstudio-a-million-ways/#using-the-venndiagram-r-package Venn Diagram with R or RStudio: A Million Ways]. Question: why the sets orders are changed? | |||
<li>Example from the help | <li>Example from the help | ||
<pre> | <pre> | ||
| Line 25: | Line 84: | ||
<li>And a [https://stackoverflow.com/a/11935618 trick] to make scaling works. </li> | <li>And a [https://stackoverflow.com/a/11935618 trick] to make scaling works. </li> | ||
</ul> | </ul> | ||
= ggvenn = | |||
* [https://cloud.r-project.org/web/packages/ggvenn/index.html ggvenn] package | |||
* [https://bioinformatics-core-shared-training.github.io/Bulk_RNAseq_Course_2021/Markdowns/11_Annotation_and_Visualisation.html RNA-seq Analysis in R] | |||
<syntaxhighlight lang='rsplus'> | <syntaxhighlight lang='rsplus'> | ||
library(ggvenn) | library(ggvenn) | ||
| Line 32: | Line 93: | ||
fill_color = c("#0073C2FF", "#CD534CFF", "#EFC000FF")) # looks normal | fill_color = c("#0073C2FF", "#CD534CFF", "#EFC000FF")) # looks normal | ||
</syntaxhighlight> | </syntaxhighlight> | ||
< | |||
< | Cons: The generated plot looks good. But the locations of set labels are weird and not adjustable. | ||
= gplots = | |||
black and white? | |||
<pre> | |||
gplots::venn(list(A = 1:150, B = 121:170, C=141:200)) | |||
</pre> | |||
= Vennerable = | |||
Vennerable package https://github.com/js229/Vennerable | |||
<ul> | <ul> | ||
<li>[https://genometoolbox.blogspot.com/2014/06/make-venn-diagram-with-correctly.html Make Venn Diagram in R with Correctly Weighted Areas] </li> | <li>[https://genometoolbox.blogspot.com/2014/06/make-venn-diagram-with-correctly.html Make Venn Diagram in R with Correctly Weighted Areas] </li> | ||
| Line 46: | Line 116: | ||
</li> | </li> | ||
</ul> | </ul> | ||
= limma = | |||
* https://rdrr.io/bioc/limma/man/venn.html | |||
* [https://bioconductor.org/packages/devel/bioc/vignettes/limma/inst/doc/usersguide.pdf User guide]. Only black and white. | |||
= eulerr = | |||
[https://cran.r-project.org/web/packages/eulerr/index.html eulerr] package | |||
= | |||
Latest revision as of 10:47, 30 April 2024
Misc
- 2014 Paper eulerAPE: Drawing Area-Proportional 3-Venn Diagrams Using Ellipses which includes an overview of several tools. The paper is linked from the website https://upset.app.
- Visualization biological data using proportional venn diagrams
- VENN DIAGRAM WITH R OR RSTUDIO: A MILLION WAYS
- http://manuals.bioinformatics.ucr.edu/home/R_BioCondManual#TOC-Venn-Diagrams and the R code or the Bioc package systemPipeR
# systemPipeR package method library(systemPipeR) setlist <- list(A=sample(letters, 18), B=sample(letters, 16), C=sample(letters, 20), D=sample(letters, 22), E=sample(letters, 18)) OLlist <- overLapper(setlist[1:3], type="vennsets") vennPlot(list(OLlist)) # R script source method source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/overLapper.R") setlist <- list(A=sample(letters, 18), B=sample(letters, 16), C=sample(letters, 20), D=sample(letters, 22), E=sample(letters, 18)) # or (obtained by dput(setlist)) setlist <- structure(list(A = c("o", "h", "u", "p", "i", "s", "a", "w", "b", "z", "n", "c", "k", "j", "y", "m", "t", "q"), B = c("h", "r", "x", "y", "b", "t", "d", "o", "m", "q", "g", "v", "c", "u", "f", "z"), C = c("b", "e", "t", "u", "s", "j", "o", "k", "d", "l", "g", "i", "w", "n", "p", "a", "y", "x", "m", "z"), D = c("f", "g", "b", "k", "j", "m", "e", "q", "i", "d", "o", "l", "c", "t", "x", "r", "s", "u", "w", "a", "z", "n"), E = c("u", "w", "o", "k", "n", "h", "p", "z", "l", "m", "r", "d", "q", "s", "x", "b", "v", "t"), F = c("o", "j", "r", "c", "l", "l", "u", "b", "f", "d", "u", "m", "y", "t", "y", "s", "a", "g", "t", "m", "x", "m" )), .Names = c("A", "B", "C", "D", "E", "F")) OLlist <- overLapper(setlist[1:3], type="vennsets") counts <- list(sapply(OLlist$Venn_List, length)) vennPlot(counts=counts)
Online tools
UpSet plot
- https://upset.app/
- UpSetR
- ComplexUpset: Create Complex UpSet Plots Using 'ggplot2' Components
- Chapter 8 UpSet plot
venn
- venn package (best so far)
- polygon() function call.
- Introduction to the venn Package in R (6 Examples) | How to Draw Up to 7 Sets
library(venn)
set.seed(12345)
x <- list(First = 1:40, Second = 15:60, Third = sample(25:50, 25),
Fourth=sample(15:65, 35))
venn(x, ilabels = "counts", zcolor = "style", ilcs = 1.2, sncs = 1)
In this example, ilcs and sncs will control the font size of the intersection labels and set names, respectively.
ggVennDiagram
https://cloud.r-project.org/web/packages/ggVennDiagram/index.html, github.
Cons: there is no way to have multiple colors for different sets.
VennDiagram
- VennDiagram. As good as the "venn" package. Many reverse imports and suggests.
- Venn Diagram with R or RStudio: A Million Ways. Question: why the sets orders are changed?
- Example from the help
grid.newpage() grid.draw(venn.diagram(list(A = 1:150, B = 121:170), NULL) ) grid.newpage() grid.draw(venn.diagram(list(A = 1:150, B = 121:170, C=141:200), NULL, category.names = c("Set 1" , "Set 2 ", "Set 3"), fill = c("#0073C2FF", "#CD534CFF", "#EFC000FF")) ) # weird - VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. scaling is one option.
- https://www.r-graph-gallery.com/venn-diagram.html
- And a trick to make scaling works.
ggvenn
- ggvenn package
- RNA-seq Analysis in R
library(ggvenn)
ggvenn(list(A = 1:150, B = 121:170, C=141:200),
fill_color = c("#0073C2FF", "#CD534CFF", "#EFC000FF")) # looks normal
Cons: The generated plot looks good. But the locations of set labels are weird and not adjustable.
gplots
black and white?
gplots::venn(list(A = 1:150, B = 121:170, C=141:200))
Vennerable
Vennerable package https://github.com/js229/Vennerable
- Make Venn Diagram in R with Correctly Weighted Areas
- Venn Diagrams with R
if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager") BiocManager::install("graph") BiocManager::install("RBGL") # fail on Rstudio cloud install.packages("Vennerable", repos="http://R-Forge.R-project.org")
limma
- https://rdrr.io/bioc/limma/man/venn.html
- User guide. Only black and white.
eulerr
eulerr package