R: Difference between revisions
| Line 279: | Line 279: | ||
</pre> | </pre> | ||
For example, the source code of ''summary'' app looks like | For example, the source code of ''summary'' app looks like | ||
<nowiki> | |||
app <- function(env) { | app <- function(env) { | ||
req <- Rook::Request$new(env) | req <- Rook::Request$new(env) | ||
res <- Rook::Response$new() | res <- Rook::Response$new() | ||
res$write('Choose a CSV file:\n') | res$write('Choose a CSV file:\n') | ||
res$write('<form method="POST" enctype="multipart/form-data">\n') | res$write('<form method="POST" enctype="multipart/form-data">\n') | ||
Revision as of 10:11, 19 November 2012
Launch Rstudio
If multiple versions of R was detected, Rstudio can not be launched successfully. A java-like clock will be spinning without a stop. The trick is to click Ctrl key and click the Rstudio at the same time. After done that, it will show up a selection of R to choose from.
Install R from source (ix86, x86_64 and arm platforms, Linux system)
Debian system (with arm architecture)
Simplest configuration
On my debian system in Pogoplug (armv5), I can compile R. See R's admin manual. If I don't need x11, I just need to install 2 required packages.
- install gfortran: apt-get install gfortran
- install readline library: apt-get install libreadline5-dev
Note: if I need x11, I should install
- libx11 and libx11-devel, libXt, libXt-devel (for fedora)
- xorg-dev (for debian)
I also run apt-get install readline-common. I don't know if this is necessary. Since I don't need x11, I use the option in configure command. After running
./configure --with-x=no --enable-R-shlib
I got
R is now configured for armv5tel-unknown-linux-gnueabi Source directory: . Installation directory: /usr/local C compiler: gcc -std=gnu99 -g -O2 Fortran 77 compiler: gfortran -g -O2 C++ compiler: g++ -g -O2 Fortran 90/95 compiler: gfortran -g -O2 Obj-C compiler: Interfaces supported: External libraries: readline Additional capabilities: NLS Options enabled: shared R library, shared BLAS, R profiling Recommended packages: yes configure: WARNING: you cannot build info or HTML versions of the R manuals configure: WARNING: you cannot build PDF versions of the R manuals configure: WARNING: you cannot build PDF versions of vignettes and help pages configure: WARNING: I could not determine a browser configure: WARNING: I could not determine a PDF viewer
However, make gave errors for recommanded packages like KernSmooth, MASS, boot, class, cluster, codetools, foreign, lattice, mgcv, nlme, nnet, rpart, spatial, and survival. The error stems from gcc: SHLIB_LIBADD: No such file or directory. Note that I can get this error message even I try install.packages("MASS", type="source").
make[1]: Entering directory `/mnt/usb/R-2.15.2/src/library/Recommended'
make[2]: Entering directory `/mnt/usb/R-2.15.2/src/library/Recommended'
begin installing recommended package MASS
* installing *source* package 'MASS' ...
** libs
make[3]: Entering directory `/tmp/Rtmp4caBfg/R.INSTALL1d1244924c77/MASS/src'
gcc -std=gnu99 -I/mnt/usb/R-2.15.2/include -DNDEBUG -I/usr/local/include -fpic -g -O2 -c MASS.c -o MASS.o
gcc -std=gnu99 -I/mnt/usb/R-2.15.2/include -DNDEBUG -I/usr/local/include -fpic -g -O2 -c lqs.c -o lqs.o
gcc -std=gnu99 -shared -L/usr/local/lib -o MASSSHLIB_EXT MASS.o lqs.o SHLIB_LIBADD -L/mnt/usb/R-2.15.2/lib -lR
gcc: SHLIB_LIBADD: No such file or directory
make[3]: *** [MASSSHLIB_EXT] Error 1
make[3]: Leaving directory `/tmp/Rtmp4caBfg/R.INSTALL1d1244924c77/MASS/src'
ERROR: compilation failed for package 'MASS'
* removing '/mnt/usb/R-2.15.2/library/MASS'
make[2]: *** [MASS.ts] Error 1
make[2]: Leaving directory `/mnt/usb/R-2.15.2/src/library/Recommended'
make[1]: *** [recommended-packages] Error 2
make[1]: Leaving directory `/mnt/usb/R-2.15.2/src/library/Recommended'
make: *** [stamp-recommended] Error 2
root@debian:/mnt/usb/R-2.15.2#
root@debian:/mnt/usb/R-2.15.2# bin/R
R version 2.15.2 (2012-10-26) -- "Trick or Treat"
Copyright (C) 2012 The R Foundation for Statistical Computing
ISBN 3-900051-07-0
Platform: armv5tel-unknown-linux-gnueabi (32-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(MASS)
Error in library(MASS) : there is no package called 'MASS'
> library()
Packages in library '/mnt/usb/R-2.15.2/library':
base The R Base Package
compiler The R Compiler Package
datasets The R Datasets Package
grDevices The R Graphics Devices and Support for Colours
and Fonts
graphics The R Graphics Package
grid The Grid Graphics Package
methods Formal Methods and Classes
parallel Support for Parallel computation in R
splines Regression Spline Functions and Classes
stats The R Stats Package
stats4 Statistical Functions using S4 Classes
tcltk Tcl/Tk Interface
tools Tools for Package Development
utils The R Utils Package
> Sys.info()["machine"]
machine
"armv5tel"
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 170369 4.6 350000 9.4 350000 9.4
Vcells 163228 1.3 905753 7.0 784148 6.0
See http://bugs.debian.org/cgi-bin/bugreport.cgi?bug=679180
Full configuration
Interfaces supported: X11, tcltk External libraries: readline Additional capabilities: PNG, JPEG, TIFF, NLS, cairo Options enabled: shared R library, shared BLAS, R profiling, Java
Install r-base and r-base-dev
In fact, if we want to take a short cut, it seems OK to run the following statement to install r-base and all required components for building r-base and extra packages. Notice that for readline package, it installs 'libreadline6-dev' instead of 'libreadline5-dev' as I just did.
Note that I did not touch /etc/apt/sources.list file so I don't know what version of R will be installed by this method.
root@debian:/mnt/usb/R-2.15.2# apt-get install r-base-dev Reading package lists... Done Building dependency tree Reading state information... Done The following package was automatically installed and is no longer required: libreadline5 Use 'apt-get autoremove' to remove them. The following extra packages will be installed: defoma dpatch file fontconfig libblas-dev libblas3gf libbz2-dev libcairo2 libdatrie1 libjpeg62-dev liblapack-dev liblapack3gf libnewt0.52 libpango1.0-0 libpango1.0-common libpaper-utils libpaper1 libpcre3-dev libpcrecpp0 libpixman-1-0 libpng12-dev libreadline-dev libreadline6-dev libthai-data libthai0 libxcb-render-util0 libxcb-render0 r-base-core unzip whiptail xdg-utils zip zlib1g-dev Suggested packages: defoma-doc psfontmgr x-ttcidfont-conf dfontmgr curl ttf-japanese-gothic ttf-japanese-mincho ttf-thryomanes ttf-baekmuk ttf-arphic-gbsn00lp ttf-arphic-bsmi00lp ttf-arphic-gkai00mp ttf-arphic-bkai00mp ess r-doc-info r-doc-pdf r-mathlib r-base-html cdbs debhelper gvfs-bin Recommended packages: libfont-freetype-perl fakeroot patchutils libfribidi0 r-recommended r-doc-html iceweasel www-browser x11-utils x11-xserver-utils shared-mime-info The following packages will be REMOVED: libreadline5-dev The following NEW packages will be installed: defoma dpatch file fontconfig libblas-dev libblas3gf libbz2-dev libcairo2 libdatrie1 libjpeg62-dev liblapack-dev liblapack3gf libnewt0.52 libpango1.0-0 libpango1.0-common libpaper-utils libpaper1 libpcre3-dev libpcrecpp0 libpixman-1-0 libpng12-dev libreadline-dev libreadline6-dev libthai-data libthai0 libxcb-render-util0 libxcb-render0 r-base-core r-base-dev unzip whiptail xdg-utils zip zlib1g-dev 0 upgraded, 34 newly installed, 1 to remove and 0 not upgraded. Need to get 22.9 MB of archives. After this operation, 58.2 MB of additional disk space will be used. Do you want to continue [Y/n]? n
Install all dependencies for building R
This is a comprehensive list. This list is even larger than r-base-dev.
root@debian:/mnt/usb/R-2.15.2# apt-get build-dep r-base Reading package lists... Done Building dependency tree Reading state information... Done The following packages will be REMOVED: libreadline5-dev The following NEW packages will be installed: bison ca-certificates ca-certificates-java debhelper defoma ed file fontconfig gettext gettext-base html2text intltool-debian java-common libaccess-bridge-java libaccess-bridge-java-jni libasound2 libasyncns0 libatk1.0-0 libaudit0 libavahi-client3 libavahi-common-data libavahi-common3 libblas-dev libblas3gf libbz2-dev libcairo2 libcairo2-dev libcroco3 libcups2 libdatrie1 libdbus-1-3 libexpat1-dev libflac8 libfontconfig1-dev libfontenc1 libfreetype6-dev libgif4 libglib2.0-dev libgtk2.0-0 libgtk2.0-common libice-dev libjpeg62-dev libkpathsea5 liblapack-dev liblapack3gf libnewt0.52 libnspr4-0d libnss3-1d libogg0 libopenjpeg2 libpango1.0-0 libpango1.0-common libpango1.0-dev libpcre3-dev libpcrecpp0 libpixman-1-0 libpixman-1-dev libpng12-dev libpoppler5 libpulse0 libreadline-dev libreadline6-dev libsm-dev libsndfile1 libthai-data libthai0 libtiff4-dev libtiffxx0c2 libunistring0 libvorbis0a libvorbisenc2 libxaw7 libxcb-render-util0 libxcb-render-util0-dev libxcb-render0 libxcb-render0-dev libxcomposite1 libxcursor1 libxdamage1 libxext-dev libxfixes3 libxfont1 libxft-dev libxi6 libxinerama1 libxkbfile1 libxmu6 libxmuu1 libxpm4 libxrandr2 libxrender-dev libxss-dev libxt-dev libxtst6 luatex m4 openjdk-6-jdk openjdk-6-jre openjdk-6-jre-headless openjdk-6-jre-lib openssl pkg-config po-debconf preview-latex-style shared-mime-info tcl8.5-dev tex-common texi2html texinfo texlive-base texlive-binaries texlive-common texlive-doc-base texlive-extra-utils texlive-fonts-recommended texlive-generic-recommended texlive-latex-base texlive-latex-extra texlive-latex-recommended texlive-pictures tk8.5-dev tzdata-java whiptail x11-xkb-utils x11proto-render-dev x11proto-scrnsaver-dev x11proto-xext-dev xauth xdg-utils xfonts-base xfonts-encodings xfonts-utils xkb-data xserver-common xvfb zlib1g-dev 0 upgraded, 136 newly installed, 1 to remove and 0 not upgraded. Need to get 139 MB of archives. After this operation, 410 MB of additional disk space will be used. Do you want to continue [Y/n]?
Web Applications
shiny
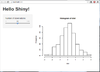
The following is what we see on a browser after we run an example from shiny package. See http://rstudio.github.com/shiny/tutorial/#hello-shiny. Note that the R session needs to be on; i.e. R command prompt will not be returned unless we press Ctrl+C or ESC.
shiny depends on websockets, caTools, bitops, digest packages.
Q & A:
- Q: If we run runExample('01_hello') in Rserve from an R client, we can continue our work in R client without losing the functionality of the GUI from shiny. Question: how do we kill the job?
- If I run the example "01_hello", the browser only shows the control but not graph on Firefox? A: Use Chrome or Opera as the default browser.
- If I run the example "01_hello" on RHEL the first time, it works fine. But if I click 'Ctrl + C' to stop it and run it again, I got a message
Warning in .SOCK_SERVE(port) : R-Websockets(tcpserv): bind() failed. Error in createContext(port, webpage, is.binary = is.binary) : Unable to bind socket on port 8100; is it realsy in use?
A: https://groups.google.com/forum/?fromgroups#!topic/shiny-discuss/jZ8S6dHFB6c
- Q: Deployment on web. A: Not ready yet. Shiny server platform is still under beta testing. Shiny apps are hosted using the R websockets package which acts more like a tcp server than a web server, and that architecture just doesn't fit with rApache, or even apache for that matter.
RApache
Rook
4 lines of code example.
library(Rook) s <- Rhttpd$new() s$start(quiet=TRUE) s$print() s$browse(1) # the cursor will return to R
We can manually open the web page http://127.0.0.1:10215/custom/RookTest.
We can add Rook application to the server; see ?Rhttpd.
s$add(
app=system.file('exampleApps/helloworld.R',package='Rook'),
name='hello'
)
s$add(
app=system.file('exampleApps/helloworldref.R',package='Rook'),
name='helloref'
)
s$add(
app=system.file('exampleApps/summary.R',package='Rook'),
name='summary'
)
s$print()
#Server started on 127.0.0.1:10221
#[1] RookTest http://127.0.0.1:10221/custom/RookTest
#[2] helloref http://127.0.0.1:10221/custom/helloref
#[3] summary http://127.0.0.1:10221/custom/summary
#[4] hello http://127.0.0.1:10221/custom/hello
# Stops the server but doesn't uninstall the app
## Not run:
s$stop()
## End(Not run)
s$remove(all=TRUE)
rm(s)
For example, the source code of summary app looks like
app <- function(env) {
req <- Rook::Request$new(env)
res <- Rook::Response$new()
res$write('Choose a CSV file:\n')
res$write('<form method="POST" enctype="multipart/form-data">\n')
res$write('<input type="file" name="data">\n')
res$write('<input type="submit" name="Upload">\n</form>\n<br>')
if (!is.null(req$POST())){
data <- req$POST()[['data']]
res$write("<h3>Summary of Data</h3>");
res$write("<pre>")
res$write(paste(capture.output(summary(read.csv(data$tempfile,stringsAsFactors=FALSE)),file=NULL),collapse='\n'))
res$write("</pre>")
res$write("<h3>First few lines (head())</h3>");
res$write("<pre>")
res$write(paste(capture.output(head(read.csv(data$tempfile,stringsAsFactors=FALSE)),file=NULL),collapse='\n'))
res$write("</pre>")
}
res$finish()
}
Stockplot
FastRWeb
Rwui
CGHWithR (removed from CRAN)
But it is still working with old version of R.
Creating local repository for CRAN and Bioconductor
How to set up a package repository
Guide: http://cran.r-project.org/doc/manuals/R-admin.html#Setting-up-a-package-repository
To create CRAN repository
Before creating a local repository please give a dry run first. You don't want to be surprised how long will it take to mirror a directory.
Dry run
rsync -avn cran.r-project.org::CRAN > crandryrun.txt
To mirror only partial repository, it is necessary to create directories before running rsync command.
mkdir /media/WD640/CRAN mkdir /media/WD640/CRAN/bin mkdir /media/WD640/CRAN/bin/windows mkdir /media/WD640/CRAN/bin/windows/contrib mkdir /media/WD640/CRAN/bin/windows/contrib/2.15 rsync -rtlzv --delete cran.r-project.org::CRAN/bin/windows/contrib/2.15/ /media/WD640/CRAN/bin/windows/contrib/2.15 (one line with space before /media) rsync -rtlzv --delete cran.r-project.org::CRAN/src/ /media/WD640/CRAN/src/
And optionally
library(tools)
write_PACKAGES("/var/www/CRAN/bin/windows/contrib/2.15", type="win.binary")
We can use du -h to check the folder size.
To create Bioconductor repository
Dry run
rsync -avn bioconductor.org::2.11 > biocdryrun.txt
Then creates directories before running rsync.
The software part (aka bioc directory) installation:
mkdir ~/Bioc/packages/2.11/bioc mkdir ~/Bioc/packages/2.11/bioc/bin mkdir ~/Bioc/packages/2.11/bioc/bin/windows mkdir ~/Bioc/packages/2.11/bioc/src rsync -zrtlv --delete bioconductor.org::2.11/bioc/bin/windows/ ~/Bioc/packages/2.11/bioc/bin/windows rsync -zrtlv --delete bioconductor.org::2.11/bioc/src/ ~/Bioc/packages/2.11/bioc/src
and annotation (aka data directory) part:
mkdir ~/Bioc/packages/2.11/data mkdir ~/Bioc/packages/2.11/data/annotation mkdir ~/Bioc/packages/2.11/data/annotation/bin mkdir ~/Bioc/packages/2.11/data/annotation/bin/windows mkdir ~/Bioc/packages/2.11/data/annotation/src rsync -zrtlv --delete bioconductor.org::2.11/data/annotation/bin/windows/ ~/Bioc/packages/2.11/data/annotation/bin/windows rsync -zrtlv --delete bioconductor.org::2.11/data/annotation/src/ ~/Bioc/packages/2.11/data/annotation/src
To test local repository
To test CRAN
r <- getOption("repos"); r["CRAN"] <- "http://arraytools.no-ip.org/CRAN"
options(repos=r)
install.packages("pamr")
To test Bioconductor
options("BioC_mirror" = "http://arraytools.no-ip.org/Bioc")
source("http://bioconductor.org/biocLite.R")
# This source biocLite.R line can be placed either before or after the previous 2 lines
biocLite("aCGH")
If there is a connection problem, check folder attributes.
chmod -R 755 ~/CRAN/bin
- Note that if a binary package was created for R 2.15.1, then it can be installed under R 2.15.1 but not R 2.15.2. The R console will show package xxx is not available (for R version 2.15.2).
- For binary installs, the function also checks for the availability of a source package on the same repository, and reports if the source package has a later version, or is available but no binary version is.
So for example, if the mirror does not have contents under src directory, we need to run the following line in order to successfully run install.packages() function.
options(install.packages.check.source = "no")
- If we only mirror the essential directories, we can run biocLite() successfully. However, the R console will give some warning
> biocLite("aCGH")
BioC_mirror: http://arraytools.no-ip.org/Bioc
Using Bioconductor version 2.11 (BiocInstaller 1.8.3), R version 2.15.
Installing package(s) 'aCGH'
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/data/experiment/src/contrib
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/extra/src/contrib
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/data/experiment/bin/windows/contrib/2.15
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/extra/bin/windows/contrib/2.15
trying URL 'http://arraytools.no-ip.org/Bioc/packages/2.11/bioc/bin/windows/contrib/2.15/aCGH_1.36.0.zip'
Content type 'application/zip' length 2431158 bytes (2.3 Mb)
opened URL
downloaded 2.3 Mb
package ‘aCGH’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\limingc\AppData\Local\Temp\Rtmp8IGGyG\downloaded_packages
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/data/experiment/bin/windows/contrib/2.15
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/extra/bin/windows/contrib/2.15
> library()
CRAN repository directory structure
The information below is specific to R 2.15.2. There are linux and macosx subdirecotries whenever there are windows subdirectory.
bin/winows/contrib/2.15 src/contrib /contrib/2.15.2 /contrib/Archive web/checks /dcmeta /packages /views
Bioconductor repository directory structure
The information below is specific to Bioc 2.11. There are linux and macosx subdirecotries whenever there are windows subdirectory.
bioc/bin/windows/contrib/2.15
/html
/install
/license
/manuals
/news
/src
/vignettes
data/annotation/bin/windows/contrib/2.15
/html
/licenses
/manuals
/src
/vignettes
/experiment/bin/windows/contrib/2.15
/html
/manuals
/src/contrib
/vignettes
extra/bin/windows/contrib
/html
/src
/vignettes
Hidden tool: rsync in Rtools
c:\Rtools\bin>rsync -avz "/cygdrive/c/users/limingc/Downloads/a.exe" "/cygdrive/c/users/limingc/Documents/" sending incremental file list a.exe sent 323142 bytes received 31 bytes 646346.00 bytes/sec total size is 1198416 speedup is 3.71 c:\Rtools\bin>
And rsync works best when we need to sync folder.
c:\Rtools\bin>rsync -avz "/cygdrive/c/users/limingc/Downloads/binary" "/cygdrive/c/users/limingc/Documents/"
sending incremental file list
binary/
binary/Eula.txt
binary/cherrytree.lnk
binary/depends64.chm
binary/depends64.dll
binary/depends64.exe
binary/mtputty.exe
binary/procexp.chm
binary/procexp.exe
binary/pscp.exe
binary/putty.exe
binary/sqlite3.exe
binary/wget.exe
sent 4115294 bytes received 244 bytes 1175868.00 bytes/sec
total size is 8036311 speedup is 1.95
c:\Rtools\bin>rm c:\users\limingc\Documents\binary\procexp.exe
cygwin warning:
MS-DOS style path detected: c:\users\limingc\Documents\binary\procexp.exe
Preferred POSIX equivalent is: /cygdrive/c/users/limingc/Documents/binary/procexp.exe
CYGWIN environment variable option "nodosfilewarning" turns off this warning.
Consult the user's guide for more details about POSIX paths:
http://cygwin.com/cygwin-ug-net/using.html#using-pathnames
c:\Rtools\bin>rsync -avz "/cygdrive/c/users/limingc/Downloads/binary" "/cygdrive/c/users/limingc/Documents/"
sending incremental file list
binary/
binary/procexp.exe
sent 1767277 bytes received 35 bytes 3534624.00 bytes/sec
total size is 8036311 speedup is 4.55
c:\Rtools\bin>
Unforunately, if the destination is a network drive, I could get a permission denied (13) error. See also http://superuser.com/questions/69620/rsync-file-permissions-on-windows
Install rgdal package on ubuntu
sudo apt-get install libgdal1-dev libproj-dev
R
> install.packages("rgdal")
Embedding R
First make sure before 'make' R, R is configured with
./configure --enable-R-shlib
Reference http://bioconductor.org/help/course-materials/2012/Seattle-Oct-2012/AdvancedR.pdf
mli@PhenomIIx6:~/Downloads/R-2.15.2/library/AdvancedR/embedding$ export R_HOME=/home/mli/Downloads/R-2.15.2
mli@PhenomIIx6:~/Downloads/R-2.15.2/library/AdvancedR/embedding$ export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/home/mli/Downloads/R-2.15.2/lib
mli@PhenomIIx6:~/Downloads/R-2.15.2/library/AdvancedR/embedding$ g++ embed.c -I/home/mli/Downloads/R-2.15.2/include -L/home/mli/Downloads/R-2.15.2/lib -lR
mli@PhenomIIx6:~/Downloads/R-2.15.2/library/AdvancedR/embedding$ R CMD ./a.out
WARNING: ignoring environment value of R_HOME
R version 2.15.2 (2012-10-26) -- "Trick or Treat"
Copyright (C) 2012 The R Foundation for Statistical Computing
ISBN 3-900051-07-0
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
ns> require(stats); require(graphics)
ns> ns(women$height, df = 5)
1 2 3 4 5
[1,] 0.000000e+00 0.000000e+00 0.00000000 0.00000000 0.0000000000
[2,] 7.592323e-03 0.000000e+00 -0.08670223 0.26010669 -0.1734044626
[3,] 6.073858e-02 0.000000e+00 -0.15030440 0.45091320 -0.3006088020
[4,] 2.047498e-01 6.073858e-05 -0.16778345 0.50335034 -0.3355668952
[5,] 4.334305e-01 1.311953e-02 -0.13244035 0.39732106 -0.2648807067
[6,] 6.256681e-01 8.084305e-02 -0.07399720 0.22199159 -0.1479943948
[7,] 6.477162e-01 2.468416e-01 -0.02616007 0.07993794 -0.0532919575
[8,] 4.791667e-01 4.791667e-01 0.01406302 0.02031093 -0.0135406187
[9,] 2.468416e-01 6.477162e-01 0.09733619 0.02286023 -0.0152401533
[10,] 8.084305e-02 6.256681e-01 0.27076826 0.06324188 -0.0405213106
[11,] 1.311953e-02 4.334305e-01 0.48059836 0.12526031 -0.0524087186
[12,] 6.073858e-05 2.047498e-01 0.59541597 0.19899261 0.0007809246
[13,] 0.000000e+00 6.073858e-02 0.50097182 0.27551020 0.1627793975
[14,] 0.000000e+00 7.592323e-03 0.22461127 0.35204082 0.4157555879
[15,] 0.000000e+00 0.000000e+00 -0.14285714 0.42857143 0.7142857143
attr(,"degree")
[1] 3
attr(,"knots")
20% 40% 60% 80%
60.8 63.6 66.4 69.2
attr(,"Boundary.knots")
[1] 58 72
attr(,"intercept")
[1] FALSE
attr(,"class")
[1] "ns" "basis" "matrix"
ns> summary(fm1 <- lm(weight ~ ns(height, df = 5), data = women))
Call:
lm(formula = weight ~ ns(height, df = 5), data = women)
Residuals:
Min 1Q Median 3Q Max
-0.38333 -0.12585 0.07083 0.15401 0.30426
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 114.7447 0.2338 490.88 < 2e-16 ***
ns(height, df = 5)1 15.9474 0.3699 43.12 9.69e-12 ***
ns(height, df = 5)2 25.1695 0.4323 58.23 6.55e-13 ***
ns(height, df = 5)3 33.2582 0.3541 93.93 8.91e-15 ***
ns(height, df = 5)4 50.7894 0.6062 83.78 2.49e-14 ***
ns(height, df = 5)5 45.0363 0.2784 161.75 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.2645 on 9 degrees of freedom
Multiple R-squared: 0.9998, Adjusted R-squared: 0.9997
F-statistic: 9609 on 5 and 9 DF, p-value: < 2.2e-16
ns> ## example of safe prediction
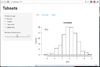
ns> plot(women, xlab = "Height (in)", ylab = "Weight (lb)")
ns> ht <- seq(57, 73, length.out = 200)
ns> lines(ht, predict(fm1, data.frame(height=ht)))
ns> ## Don't show:
ns> ## Consistency:
ns> x <- c(1:3,5:6)
ns> stopifnot(identical(ns(x), ns(x, df = 1)),
ns+ identical(ns(x, df=2), ns(x, df=2, knots=NULL)),# not true till 2.15.2
ns+ !is.null(kk <- attr(ns(x), "knots")),# not true till 1.5.1
ns+ length(kk) == 0)
ns> ## End Don't show
ns>
ns>
ns>
The above result can be compared with running
mli@PhenomIIx6:~/Downloads/R-2.15.2/library/AdvancedR/embedding$ R WARNING: ignoring environment value of R_HOME
R version 2.15.2 (2012-10-26) -- "Trick or Treat" Copyright (C) 2012 The R Foundation for Statistical Computing ISBN 3-900051-07-0 Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY. You are welcome to redistribute it under certain conditions. Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors. Type 'contributors()' for more information and 'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or 'help.start()' for an HTML browser interface to help. Type 'q()' to quit R.
> library(splines) > example("ns")
ns> require(stats); require(graphics)
ns> ns(women$height, df = 5)
1 2 3 4 5 [1,] 0.000000e+00 0.000000e+00 0.00000000 0.00000000 0.0000000000 [2,] 7.592323e-03 0.000000e+00 -0.08670223 0.26010669 -0.1734044626 [3,] 6.073858e-02 0.000000e+00 -0.15030440 0.45091320 -0.3006088020 [4,] 2.047498e-01 6.073858e-05 -0.16778345 0.50335034 -0.3355668952 [5,] 4.334305e-01 1.311953e-02 -0.13244035 0.39732106 -0.2648807067 [6,] 6.256681e-01 8.084305e-02 -0.07399720 0.22199159 -0.1479943948 [7,] 6.477162e-01 2.468416e-01 -0.02616007 0.07993794 -0.0532919575 [8,] 4.791667e-01 4.791667e-01 0.01406302 0.02031093 -0.0135406187 [9,] 2.468416e-01 6.477162e-01 0.09733619 0.02286023 -0.0152401533
[10,] 8.084305e-02 6.256681e-01 0.27076826 0.06324188 -0.0405213106 [11,] 1.311953e-02 4.334305e-01 0.48059836 0.12526031 -0.0524087186 [12,] 6.073858e-05 2.047498e-01 0.59541597 0.19899261 0.0007809246 [13,] 0.000000e+00 6.073858e-02 0.50097182 0.27551020 0.1627793975 [14,] 0.000000e+00 7.592323e-03 0.22461127 0.35204082 0.4157555879 [15,] 0.000000e+00 0.000000e+00 -0.14285714 0.42857143 0.7142857143 attr(,"degree") [1] 3 attr(,"knots")
20% 40% 60% 80%
60.8 63.6 66.4 69.2 attr(,"Boundary.knots") [1] 58 72 attr(,"intercept") [1] FALSE attr(,"class") [1] "ns" "basis" "matrix"
ns> summary(fm1 <- lm(weight ~ ns(height, df = 5), data = women))
Call: lm(formula = weight ~ ns(height, df = 5), data = women)
Residuals:
Min 1Q Median 3Q Max
-0.38333 -0.12585 0.07083 0.15401 0.30426
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 114.7447 0.2338 490.88 < 2e-16 *** ns(height, df = 5)1 15.9474 0.3699 43.12 9.69e-12 *** ns(height, df = 5)2 25.1695 0.4323 58.23 6.55e-13 *** ns(height, df = 5)3 33.2582 0.3541 93.93 8.91e-15 *** ns(height, df = 5)4 50.7894 0.6062 83.78 2.49e-14 *** ns(height, df = 5)5 45.0363 0.2784 161.75 < 2e-16 *** --- Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Residual standard error: 0.2645 on 9 degrees of freedom Multiple R-squared: 0.9998, Adjusted R-squared: 0.9997 F-statistic: 9609 on 5 and 9 DF, p-value: < 2.2e-16
ns> ## example of safe prediction
ns> plot(women, xlab = "Height (in)", ylab = "Weight (lb)")
ns> ht <- seq(57, 73, length.out = 200)
ns> lines(ht, predict(fm1, data.frame(height=ht)))
ns> ## Don't show: ns> ## Consistency: ns> x <- c(1:3,5:6)
ns> stopifnot(identical(ns(x), ns(x, df = 1)), ns+ identical(ns(x, df=2), ns(x, df=2, knots=NULL)),# not true till 2.15.2 ns+ !is.null(kk <- attr(ns(x), "knots")),# not true till 1.5.1 ns+ length(kk) == 0)
ns> ## End Don't show ns> ns> ns>
Note that if I follow the instruction to put embed.c at the end of g++ command, I will get an error.
mli@PhenomIIx6:~/Downloads/R-2.15.2/library/AdvancedR/embedding$ g++ -I/home/mli/Downloads/R-2.15.2/include -L/home/mli/Downloads/R-2.15.2/lib -lR embed.c /tmp/cc7Vum5j.o: In function `main': embed.c:(.text+0x1c): undefined reference to `Rf_initEmbeddedR' embed.c:(.text+0x2b): undefined reference to `Rf_endEmbeddedR' /tmp/cc7Vum5j.o: In function `doSplinesExample()': embed.c:(.text+0x45): undefined reference to `Rf_mkString' embed.c:(.text+0x52): undefined reference to `Rf_install' embed.c:(.text+0x5d): undefined reference to `Rf_lang2' embed.c:(.text+0x6d): undefined reference to `Rf_protect' embed.c:(.text+0x74): undefined reference to `R_GlobalEnv' embed.c:(.text+0x87): undefined reference to `R_tryEval' embed.c:(.text+0x91): undefined reference to `Rf_unprotect' embed.c:(.text+0x9b): undefined reference to `Rf_ScalarLogical' embed.c:(.text+0xa8): undefined reference to `Rf_install' embed.c:(.text+0xb3): undefined reference to `Rf_lang2' embed.c:(.text+0xc3): undefined reference to `Rf_protect' embed.c:(.text+0xcd): undefined reference to `Rf_install' embed.c:(.text+0xdc): undefined reference to `CDR' embed.c:(.text+0xe7): undefined reference to `SET_TAG' embed.c:(.text+0xee): undefined reference to `R_GlobalEnv' embed.c:(.text+0x102): undefined reference to `R_tryEval' embed.c:(.text+0x10c): undefined reference to `Rf_unprotect' embed.c:(.text+0x116): undefined reference to `Rf_mkString' embed.c:(.text+0x123): undefined reference to `Rf_install' embed.c:(.text+0x12e): undefined reference to `Rf_lang2' embed.c:(.text+0x13e): undefined reference to `Rf_protect' embed.c:(.text+0x145): undefined reference to `R_GlobalEnv' embed.c:(.text+0x158): undefined reference to `R_tryEval' embed.c:(.text+0x162): undefined reference to `Rf_unprotect' collect2: ld returned 1 exit status