Biowulf: Difference between revisions
| Line 139: | Line 139: | ||
b1 5/20 300/640 16 32 250g 400g cpu32,core16,g256,sata400,e2670,1g | b1 5/20 300/640 16 32 250g 400g cpu32,core16,g256,sata400,e2670,1g | ||
b1 3/10 100/160 8 16 68g 100g cpu16,core8,g72,sata100,x5550,1g | b1 3/10 100/160 8 16 68g 100g cpu16,core8,g72,sata100,x5550,1g | ||
</syntaxhighlight> | |||
== Running R scripts == | |||
https://hpc.nih.gov/apps/R.html | |||
Running a swarm of R batch jobs on Biowulf | |||
<syntaxhighlight lang='bash'> | |||
R --vanilla < /data/username/R/R1 > /data/username/R/R1.out | |||
R --vanilla < /data/username/R/R2 > /data/username/R/R2.out | |||
</syntaxhighlight> | |||
swarm -g 16 -f /home/username/Rjobs --module R | |||
Parallelizing with 'parallel' | |||
<syntaxhighlight lang='bash'> | |||
detectBatchCPUs <- function() { | |||
ncores <- as.integer(Sys.getenv("SLURM_CPUS_PER_TASK")) | |||
if (is.na(ncores)) { | |||
ncores <- as.integer(Sys.getenv("SLURM_JOB_CPUS_PER_NODE")) | |||
} | |||
if (is.na(ncores)) { | |||
return(4) # for helix | |||
} | |||
return(ncores) | |||
} | |||
ncpus <- detectBatchCPUs() | |||
options(mc.cores = ncpus) | |||
mclapply(..., mc.cores = ncpus) | |||
makeCluster(ncpus) | |||
</syntaxhighlight> | </syntaxhighlight> | ||
Revision as of 14:11, 17 November 2016
- Biowulf User Guide. Note that Biowulf2 runs Centos (RedHat) 6.x. (Biowulf1 is at Centos 5.x)
Linux distribution
$ ls /etc/*release # login mode $ cat /etc/redhat-release Red Hat Enterprise Linux Server release 6.8 (Santiago) $ sinteractive # switch to biowulf2 computing nodes $ cat /etc/redhat-release CentOS release 6.6 (Final) $ cat /etc/centos-release CentOS release 6.6 (Final)
Training notes
- https://hpc.nih.gov/docs/
- https://hpc.nih.gov/docs/B2training.pdf
- https://hpc.nih.gov/docs/biowulf2-beta-handout.pdf
Storage
Quota
checkquota
Environment modules
# What modules are available
module avail
module -d avail # default
module avail STAR
module spider bed # search by a case-insensitive keyword
# Load a module
module list # loaded modules
module load STAR
module load STAR/2.4.1a
module load plinkseq macs bowtie # load multiple modules
# Unload a module
module unload STAR/2.4.1a
# Switch to a different version of an application
# If you load a module, then load another version of the same module, the first one will be unloaded.
# Examine a modulefile
$ module display STAR
-----------------------------------------------------------------------------
/usr/local/lmod/modulefiles/STAR/2.5.1b.lua:
-----------------------------------------------------------------------------
help([[This module sets up the environment for using STAR.
Index files can be found in /fdb/STAR
]])
whatis("STAR: ultrafast universal RNA-seq aligner")
whatis("Version: 2.5.1b")
prepend_path("PATH","/usr/local/apps/STAR/2.5.1b/bin")
# Set up personal modulefiles
# Using modules in scripts
# Shared Modules
Single file - sbatch
- sbatch
- Note sbatch command does not support --module option. In sbatch case, the module command has to be put in the script file.
- Script file must be starting with a line #!/bin/bash
Don't use the swarm command on a single script file since swarm will treat each line of the script file as an independent command.
sbatch --cpus-per-task=2 --mem=4g MYSCRIPT
An example of the script file (Slurm environment variable $SLURM_CPUS_PER_TASK within your script was used to specify the number of threads to the program)
#!/bin/bash module load novocraft novoalign -c $SLURM_CPUS_PER_TASK -f s_1_sequence.txt -d celegans -o SAM > out.sam
Multiple files - swarm
swarm -t 3 -g 20 -f run_seqtools_vc.sh --module samtools,picard,bwa --verbose 1 --devel # 3 commands run in 3 subjobs, each command requiring 20 gb and 3 threads, allocating 6 cores and 12 cpus swarm -t 3 -g 20 -f run_seqtools_vc.sh --module samtools,picard,bwa --verbose 1 # To change the default walltime, use --time=24:00:00 swarm -t 8 -g 24 --module tophat,samtools,htseq -f run_master.sh cat sw3n17156.o
Partition and freen
- https://hpc.nih.gov/docs/b2-userguide.html#cluster_status
- In the script file, we can use $SLURM_CPUS_PER_TASK to represent the number of cpus
- In the swarm command, we can use -t to specify the threads and -g to specify the memory (GB).
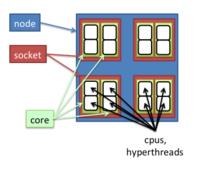
Biowulf nodes are grouped into partitions. A partition can be specified when submitting a job. The default partition is 'norm'. The freen command can be used to see free nodes and CPUs, and available types of nodes on each partition.
We may need to run swarm commands on non-default partitions. For example, not many free CPUs are available in 'norm' partition. Or Total time for bundled commands is greater than partition walltime limit. Or because the default partition norm has nodes with a maximum of 120GB memory.
We can run the swarm command on different partition (the default is 'norm'). For example, to run on b1 parition (the hardware in b1 looks inferior to norm)
swarm -f outputs/run_seqtools_dge_align -g 20 -t 16 --module tophat,samtools,htseq --time=6:00:00 --partition b1 --verbose 1
Below is an output from freen command.
........Per-Node Resources........ Partition FreeNds FreeCPUs Cores CPUs Mem Disk Features -------------------------------------------------------------------------------- norm* 0/301 1624/9488 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,10g unlimited 3/12 202/384 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,10g niddk 1/82 228/2624 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,10g,niddk ibfdr 0/184 0/5888 16 32 60g 800g cpu32,core16,g64,ssd800,x2650,ibfdr ibqdr 1/95 32/3040 16 32 29g 400g cpu32,core16,g32,sata400,x2600,ibqdr ibqdr 51/89 1632/2848 16 32 60g 400g cpu32,core16,g64,sata400,x2600,ibqdr gpu 1/4 116/128 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,gpuk20x,acemd gpu 17/20 634/640 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,gpuk20x largemem 3/4 254/256 32 64 1007g 800g cpu64,core32,g1024,ssd800,x4620,10g nimh 58/60 1856/1920 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,10g,nimh ccr 0/85 1540/2720 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,10g,ccr ccr 0/63 598/2016 16 32 123g 400g cpu32,core16,g128,sata400,x2600,1g,ccr ccr 0/60 974/1920 16 32 60g 400g cpu32,core16,g64,sata400,x2600,1g,ccr ccrclin 4/4 128/128 16 32 60g 400g cpu32,core16,g64,sata400,x2600,1g,ccr quick 0/85 1540/2720 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,10g,ccr quick 0/63 598/2016 16 32 123g 400g cpu32,core16,g128,sata400,x2600,1g,ccr quick 0/60 974/1920 16 32 60g 400g cpu32,core16,g64,sata400,x2600,1g,ccr quick 1/82 228/2624 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,10g,niddk quick 58/60 1856/1920 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,10g,nimh b1 71/363 7584/8712 12 24 21g 100g cpu24,core12,g24,sata100,x5660,1g b1 3/287 3410/4592 8 16 21g 200g cpu16,core8,g24,sata200,x5550,1g b1 7/16 192/256 8 16 68g 200g cpu16,core8,g72,sata200,x5550,1g b1 5/20 300/640 16 32 250g 400g cpu32,core16,g256,sata400,e2670,1g b1 3/10 100/160 8 16 68g 100g cpu16,core8,g72,sata100,x5550,1g
Running R scripts
https://hpc.nih.gov/apps/R.html
Running a swarm of R batch jobs on Biowulf
R --vanilla < /data/username/R/R1 > /data/username/R/R1.out R --vanilla < /data/username/R/R2 > /data/username/R/R2.out
swarm -g 16 -f /home/username/Rjobs --module R
Parallelizing with 'parallel'
detectBatchCPUs <- function() {
ncores <- as.integer(Sys.getenv("SLURM_CPUS_PER_TASK"))
if (is.na(ncores)) {
ncores <- as.integer(Sys.getenv("SLURM_JOB_CPUS_PER_NODE"))
}
if (is.na(ncores)) {
return(4) # for helix
}
return(ncores)
}
ncpus <- detectBatchCPUs()
options(mc.cores = ncpus)
mclapply(..., mc.cores = ncpus)
makeCluster(ncpus)
Monitor jobs/Delete jobs
https://hpc.nih.gov/docs/userguide.html#monitor
sjobs watch -n 30 jobload scancel -u XXXXX scancel NNNNN scancel --state=PENDING scancel --state=RUNNING squeue -u XXXX jobhist 17500 # report the CPU and memory usage of completed jobs.
The other two commands are very useful too jobhist and swarmhist (temporary).
$ cat /usr/local/bin/swarmhist
#!/bin/bash
usage="usage: $0 jobid"
jobid=$1
[[ -n $jobid ]] || { echo $usage; exit 1; }
ret=$(grep "jobid=$jobid" /usr/local/logs/swarm_on_slurm.log)
[[ -n $ret ]] || { echo "no swarm found for jobid = $jobid"; exit; }
echo $ret | tr ';' '\n'
$ jobhist 22038972
$ swarmhist 22038972
date=(SKIP)
host=(SKIP)
jobid=22038972
user=(SKIP)
pwd=(SKIP)
ncmds=1
soptions=--array=0-0 --job-name=swarm --output(SKIP)
njobs=1
job-name=swarm
command=/usr/local/bin/swarm -t 16 -g 20 -f outputs/run_seqtools_vc --module samtools,picard --verbose 1
Exit code
https://hpc.nih.gov/docs/b2-userguide.html#exitcodes
Local disk and temporary files
https://hpc.nih.gov/docs/b2-userguide.html#local
Interactive debugging
8GB and 4 CPUs on a single node. Increase them to 60 GB and more cores if we run something like STAR for rna-seq reads alignment.
sinteractive --mem=8g -c 4
Parallel jobs
Parallel (MPI) jobs that run on more than 1 node: Use the environment variable $SLURM_NTASKS within the script to specify the number of MPI processes.