Rstudio
Different tastes
Server
Like the Cloud version (a little better than the cloud version), the connection is unstable:(
So it is unusable.
Cloud
https://rstudio.cloud/. You can sign up using gmail or github account.
Free account limitation: RStudio Cloud cannot read in 1GB file? It seems 1GB memory limit still holds as of today (2020-03-09). Use cat /sys/fs/cgroup/memory/memory.limit_in_bytes to find out the limit (1073741824 in this case).
Upload a file: Click 'Files' under the bottom right pane (Files-Plots-Packages-Help-Viewer). There is an "upload" button next to the "New folder" button over there.
I can install a package from a source with URL. So I don't need to upload the packages to RStudio.
If it took too long to load an existing project (such as shiny projects), I just need to relaunch the project (Click three dots icon and select the 3rd option).
I got an error "Unexpected response from server" when I try to upload a file around 300MB. 2020-02-26.
I got an error "Error occurred during transmission" when I try to run an R statement.
Desktop
Build from source See https://github.com/rstudio/rstudio/blob/master/INSTALL for installation instruction.
To build Rstudio from source in Ubuntu:
- Install R.
- Install Qt 4.
- Download a release version tarball of source code. Do not grab from github master branch. Sometimes the preview version works but the release version cannot be compiled. For example, the release version 0.98.507 would give a build error from /opt/QRstudio-QtSDK/Desktop/4.8.0/gcc/include/QtCore/gstring.h, but the preview version 0.98.836 (May 11th, 2014) works fine.
- Make sure git is installed/available.
- Install dependencies (note that installing boost will take a few minutes. Make sure there are 2GB memory or will get a memory related error when running make install)
cd rstudio-rstudio-dbf531d # the number at the end depending on the source cd dependencies/linux ./install-dependencies-debian
- Create build directory
mkdir build cd build
- Configure by running cmake
cmake .. -DRSTUDIO_TARGET=Desktop -DCMAKE_BUILD_TYPE=Release
If something is wrong related to boost library, go back to step 1. Then type (installing boost library takes a while)
cd dependencies/linux run ./install-boost
And for some reason, when I try to run cmake (OS is 32-bit LXLE running on VM), I get the following error
CMake Error at /usr/share/cmake-2.8/Modules/FindPackageHandleStandardArgs.cmake:108 (message): Could NOT find Qt4 (missing: QT_QTWEBKIT_INCLUDE_DIR QT_QTWEBKIT_LIBRARY QT_QTDBUS_INCLUDE DIR QT_QTDBUS_LIBRARY) (found suitable version "4.8.6", minimum required is "4.8.0")
- Build and Install
sudo make install
For RStudio Desktop on Linux, make install automatically creates an entry in the Applications -> Programming menu for RStudio. The rstudio can be launched by /usr/local/lib/rstudio/bin/rstudio.
Releases
- RStudio v1.4 Preview: Visual Markdown Editing, Python Support, Command Palette, Citations, The Little Things
- RStudio 1.3 Preview: Integrated Tutorials: learnr package. learnr 0.10.0.
- 1.2
Discussion
Preferences
RStudio Theme
Background color (v1.1.383, see an animation on this post)
There are 3 choices of RStudio theme (Using RStudio Themes):
- Classic + selected Editor theme: Code and Console/Terminal are dark, Environment/history/connections and Files/Plots/Packages/Help/Viewer are white (good when doing finding). But the 'R Markdown' tab does not show a working rotation symbol.
- Modern: 4 panels are all dark (my choice).
- Sky: 4 panels are all dark
One good-looking option is also change the layout like this one. So source code take half and console and plot each takes a quarter of the screen.
Background color of plot pane becomes black. It turns out the reason is the user is using a transparent background in his plotting routine.
Custom Theme support was added in RStudio v1.2.
Editor theme
I pick "Vibrant Ink".
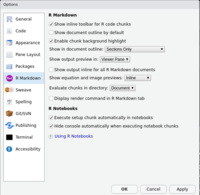
R markdown
- Rstudio - How to show plot output in bottom right pane?
- View markdown generated html in RStudio viewer
Toolbar
View -> Hide Toolbar
Change repository
- chooseCRANmirror(): this actually changes the CRAN mirror. It is done mainly interactively.
- Non-interactive method.
local({ r <- getOption("repos") r["CRAN"] <- "https://mirror.las.iastate.edu/CRAN" options(repos = r) }) - setRepositories(ind=1:2) (both non-interactive or interactive): this allows to install packages from Bioconductor. After the change, we can use RStudio or the command install.packages() to install R package from Bioconductor. See setRepositories documentation.
1: + CRAN 2: + BioC software 3: BioC annotation 4: BioC experiment 5: CRAN (extras) 6: Omegahat 7: R-Forge 8: rforge.net
In my case, stringr package source code in cloud.r-project.org (default in RStudio or 0-Cloud from chooseCRANmirror()) is 1 week old. That version of source code is now archived and the URL shown from the output of install.packages() cannot be found. When I switch to USA-IA mirror, it has the latest version.
Use options("repos") to see its effect.
options("repos")
$`repos`
CRAN BioCsoft
"https://mirror.las.iastate.edu/CRAN" "https://bioconductor.org/packages/3.7/bioc"
Change R version
Desktop
https://support.rstudio.com/hc/en-us/articles/200486138-Changing-R-versions-for-RStudio-desktop
On Linux, the default R can be found by which R in the command line. If you want to override which version of R is used then you can set the RSTUDIO_WHICH_R environment variable to the R executable that you want to run against. For example:
export RSTUDIO_WHICH_R=/opt/R/3.5.0/bin/R
Not that in order for RStudio to see this environment variable when launched from the Ubuntu desktop Applications menu (as opposed to from a terminal) it must be defined in the ~/.profile file.
Server
Using multiple versions of R with RStudio Server Pro
pandoc
- https://pandoc.org/installing.html
- How to make R Markdown use an updated version of pandoc on my Mac
$ pandoc --version pandoc 2.7.3 $ /Applications/RStudio.app/Contents/MacOS/pandoc/pandoc -v pandoc 2.3.1
To use the system's pandoc, create/modify project's .Renviron file and include a line to something like
PATH="/usr/local/bin/:${PATH}"
Get help on firefox/chrome browser
(The help panel is kind of small)
No, it can't. But alternatively, if we click "Show in a new window" icon, it will bring the help on an independent window.
> getOption("browser")
function (url)
{
.Call("rs_browseURL", url)
}
<environment: 0x1121bda60>
Tips
Outline
Click a small icon/button at the top-right corner of the code panel.
- For an Rmd file, it gives an online (all section headers) of the file.
- For an R file, it list all functions in the document.
Some good examples from rstudio.
Install package
(Mac OS). If I install a package in R console, the package will be installed under '/Users/XXX/Library/R/3.6/library'.
But if I install the package using RStudio's packages panel or through 'menu > Tools > Install Packages...', the package will go to "/Library/Frameworks/R.framework/Versions/3.6/Resources/library". So if we use remove.packages() without specify the lib parameter, it may there is no package that can be uninstalled. We can change the installation directory if we install the package using 'menu > Tools > Install Packages...' method. Here has some screenshots on Windows OS.
Pop out the source pane to have its own window
Drag the tab outside the RStudio window to where you want your new code window to appear, or Click the "Show in new window" button on the editor toolbar (it's next to forward/back)
However, it seems we cannot move the popped out pane back to RStudio. If we just want to maximize one pane, we can use the keyboard shortcut "Ctrl+Shift+1" (source pane) or "Ctrl+Shift+2" (console pane); see Zoom in one pane.
Column selection
Click Alt.
Menu -> Code
- For R code files *.R, it has "Insert Section" (a recommended way to insert a new section)
- For R markdown files *.Rmd, it has "Insert Chunk" (just a short cut)
Restart R
Three ways
- Session -> Restart R
- .rs.restart()
- command/ctrl + shift + F10
Note Restarting R in RStudio won't delete/clear workspace/objects but it unloads R packages.
We can use Session -> Clear Workspace or the command rm(list=ls()) to delete all objects.
debug location is approximate because the source is not available
- https://stackoverflow.com/a/31662904
- https://stackoverflow.com/a/50268331. I have tried something like (it does not help)
install.packages("survminer", INSTALL_opts = "--with-keep.source --no-byte-compile") debug(survminer::ggsurvplot) ggsurvplot(...)
Using the RStudio Terminal
https://support.rstudio.com/hc/en-us/articles/115010737148-Using-the-RStudio-Terminal
On my Mac, I need to run chsh -s /bin/bash AND then source ~/.bashrc in order to enable the changes I want.
Why doesn't .bashrc run automatically?
Snippet
- https://support.rstudio.com/hc/en-us/articles/204463668-Code-Snippets?mobile_site=true
- If you type 'ts' into the console, it should pop up a list. However, the default completion delay is 3 characters, meaning it will not work unless your completion delay is set to 2 characters which you can change in your global options (Code > Completion). This works on R console or an R/Rmd file.
- If you don't want to change your default delay you can just type ts and hit tab.
Crash and log file
On my Mac, the crash report is saved at ~/Library/Logs/DiagnosticReports/rsession_XXXX.crash and the log file is saved at ~/.rstudio-desktop/log/.
From within RStudio: Help Menu -> Diagnostics -> Show Log Files
See also RStudio Crash Report, RStudio Application Logs.
Check if R is running in RStudio
Sys.getenv("RSTUDIO") == "1"
# Or
.Platform$GUI == "RStudio"
Install on Crouton
Test on my Asus (intel CPU) Chromebox/Crouton with Ubuntu 14.04.
(trusty)brb@localhost:~$ sudo dpkg -i '/tmp/mozilla_brb0/rstudio-1.0.44-amd64.deb' Selecting previously unselected package rstudio. (Reading database ... 133157 files and directories currently installed.) Preparing to unpack .../rstudio-1.0.44-amd64.deb ... Unpacking rstudio (1.0.44) ... dpkg: dependency problems prevent configuration of rstudio: rstudio depends on libjpeg62; however: Package libjpeg62 is not installed. dpkg: error processing package rstudio (--install): dependency problems - leaving unconfigured Processing triggers for shared-mime-info (1.2-0ubuntu3) ... Processing triggers for hicolor-icon-theme (0.13-1) ... Processing triggers for gnome-menus (3.10.1-0ubuntu2) ... Processing triggers for desktop-file-utils (0.22-1ubuntu1) ... Processing triggers for bamfdaemon (0.5.1+14.04.20140409-0ubuntu1) ... Rebuilding /usr/share/applications/bamf-2.index... Processing triggers for mime-support (3.54ubuntu1.1) ... Errors were encountered while processing: rstudio
After I run sudo apt-get install libjpeg62, RStudio can be installed successfully.
Switch R version for RStudio desktop
https://support.rstudio.com/hc/en-us/articles/200486138-Changing-R-versions-for-RStudio-desktop
In Linux desktop, we need to run the following (or include the line in ~/.profile file for a permanent solution) before calling 'rstudio' in a terminal.
export RSTUDIO_WHICH_R=/opt/R/3.5.0/bin/R
Stop a command
- Use the stop sign
- Use "Interrupt R" under Session menu.
See Kill a process in R with RStudio
Mouse
Scroll different tabs using the mouse wheel after you click any one of tabs.
Keyboard shortcut
- https://support.rstudio.com/hc/en-us/articles/200711853-Keyboard-Shortcuts (Help -> Keyboard Shortcut Quick Reference)
- Customize the shortcuts.
- https://www.dropbox.com/s/0moch6xocili6v9/RStudio%20Shortcuts.pdf?dl=0
- Send shell script (*.sh) code line by line will be done in Terminal tab. If we click 'Run Script', it will call system() in R.
- Knit an rmarkdown file
- Mac: Shift + Cmd + k
- Linux: Shift + Ctrl + k
- (Aug 16, 2018) for some reason, run current line (CMD+Enter) will send random lines. It does not help if I assign a different key-bind. Update RStudio does not help either.
- One way it still works is by selecting line(s) first and hitting the key-bind.
- Another way is to go to Tools > Global options > Code. Change to Current line from the dropdown list next to CMD + Enter executes. The default is "Multiple-line R statements" which should be better since an R statement can spread several lines. The 3rd option "Multiple consecutive R lines" will send a block of R lines automatically which does not look ideal in most cases.
Move focus to source/console
- Ctrl + 1: source
- Ctrl + 2: console
These are the default.
Zoom in/out on one pane/window
- Ctrl + Shift + 1: source
- Ctrl + Shift + 2: console
- Ctrl + Shift + 3: file viewer
- Ctrl + Shift + 4: environment
- Ctrl + Shift + 0: show all panes
Show previous/next Plot
By default, they are "Ctrl + Alt + F11" and "Ctrl + Alt + F12". But they have a conflict with other shortcuts in RStudio IDE itself or with the (Linux/Mint) system. In my case, Ctrl+Alt+F11 will escape from Linux Desktop:( Also the Quick Reference shows these bindings had a conflict: Ctrl + Alt + F11 represents Previous Terminal or Previous Plot.
I change the shortcuts to Alt+Left and Alt+Right (directly use keyboard to change it). It works but there is a delay if the plot is more complicated (6 seconds on a 10000 genes heatmap). These new assignment works on Mac and Linux.
- Cmd + up/down arrow keys (Mac)
- Ctrl + up/down arrow keys (Linux)
Viewer pane
- Make R's View() open in a new window automatically
- Is it possible to view an HTML table in the viewer pane?
View() output
Save View() output of RStudio as html
Automatically reformat code
- CMD + Shift + A (mac)
- Ctrl + Shift + A (Windows)
Fix code spacing, reindent lines
CMD + i
History panel
If we want to rerun lines from the history panel, just highlight the lines and then either click the "To Console" button or press the "Enter" key.
See Using Command History from RStudio.
RStudio server
KVM
Multiple Rstudio Server instances using a single R/LaTeX install with KVM
RStudio Server Installation on Ubuntu
https://rviews.rstudio.com/2017/06/21/analytics-administration-for-r/
RStudio Server on Amazon EC2
RStudio and Git
- Using Version Control with RStudio. The link is available from RStudio -> Tools -> Project Options -> Git/SVN. ? Using Version Control with RStudio
- Happy Git and GitHub for the useR Jenny Bryan
- http://www.datasurg.net/2015/07/13/rstudio-and-github/
- Git window on RStudio IDE http://r-pkgs.had.co.nz/git.html
Plots window
- The plots window (device) can only show 1 plot (even I use zoom to open another window).
- If I use dev.off(), it will erase all plots in the plots window (tested on Mac). But if I have multiple graphical devices (see next), I should use dev.off(dev.list()["RStudioGD"]). See Code to clear all plots in RStudio.
- In order to open another window, use x11(), windows() or quartz() (depending on your device) before each plot. Then call dev.off() to clear last plot. See How do you delete the current (but not all) plots in the RStudio plotting device?
Plot does not show up in RStudio's plot window
options(device = "RStudioGD") # or dev.off() dev.new() ...
Overlap problem
https://stackoverflow.com/questions/45395220/legends-overlapping-the-plots-in-rstudio
Project
Using Projects
Working with Multiple Projects at Once
Sharing a Project with multiple users
https://support.rstudio.com/hc/en-us/articles/201751487-Sharing-a-project-with-multiple-users
Project settings
Menu -> Tools -> Project Options... The changes are saved in XXX.Rproj file.
.Rproj.user directory
We probably don't want to share this directory with others.
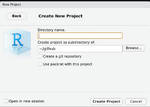
This directory is also automatically added to .Rbuildignore, .gitignore, etc. if we check the option Create a git repository when creating a new project.
Note when we create a new project, we can check the option Open in a new session (default is not checked).
Script for creating a new project in Rstudio
https://stackoverflow.com/a/26127716. Below is a modified with my fix. It will generate a new file XXX.Rproj under the current working directory.
myProject <- function(proj, ...) {
#require(ProjectTemplate)
#create.project(proj, ...)
x <- c("Version: 1.0", "", "RestoreWorkspace: Default", "SaveWorkspace: Default",
"AlwaysSaveHistory: Default", "", "EnableCodeIndexing: Yes",
"UseSpacesForTab: Yes", "NumSpacesForTab: 4", "Encoding: UTF-8",
"", "RnwWeave: knitr", "LaTeX: pdfLaTeX")
#cat(paste(x, collapse="\n"), file=file.path(proj, paste0(basename(proj), ".Rproj")))
cat(paste(x, collapse="\n"), file=paste0(basename(proj), ".Rproj"))
message(paste0(basename(proj), ".Rproj has been created"))
}
myProject("MyNewProject") # It will generate the file <MyNewProject.Rproj>.
Running A Data Project In R
https://kkulma.github.io/2018-03-18-Prime-Hints-for-Running-a-data-project-in-R/
- Use R projects. Always
- Describe the purpose of your code / project.
- Load all necessary packages in the beginning of your script & consider calling functions in a package via ::
- Name your code sections and use them for quick navigation
- Make your life easier and mark your code when you need to
- Write your code as if someone was going to use it without communicating with you. From. Day. One
- Name your files like a Pro: Machine readable, human readable & play well with ordering
- Put something numeric in your file name first.
- use the YYYY-MM-DD format for dates
- regular expression and globbing friendly
- easy to compute on using delimiters
- left pad other numbers with zeroes
- With big, complex data projects use project pipeline
- load.R
- clean.R
- func.R
- do.R
- Never save your workspace
- Before publishing/sharing your code, run it in the fresh workspace
- https://stackoverflow.com/questions/49549962/how-to-give-a-title-for-a-block-of-r-codes-denoted-by-five-or-more-e-x
- Code Folding and Sections and Navigating Code from RStudio
- Hadley's Commenting guidelines. Note that the object documentation lines won't show up in the navigation section.
- Function definitions do not form a code section though the functions are shown in the R script function menu (this term is borrowed from Navigating Code) post. Function definitions are nested under sections.
The best practice to include a comment for a block of code is
- Code > Insert Section (this will create something like # Section One --------------------------------- )
- At least one number sign (pound sign) before the title PLUS at least 4 of number signs/dash signs/equal signs at the end
Good examples
#### My section #### # My section #### # My section ----
Bad example 1 (this will show 2 lines of (Untitled) in RStudio nagivation)
################ # Section 1 ################
Bad example 2 (this will not show anything in the navigation)
# Section 2 # Section 2 # # Section 2 ## # Section 2 ###
Jobs
- Detailed progress in running a job in RStudio.
- How to run R scripts as RStudio local background jobs
Python
R and Python: How to Integrate the Best of Both into Your Data Science Workflow
Addins
- https://rstudio.github.io/rstudioaddins/
- https://support.rstudio.com/hc/en-us/articles/215605467
- https://resources.rstudio.com/rstudio-conf-2019/empowering-a-data-team-with-rstudio-addins (Video)
Step-by-step to create an addin R package
Defining Your Own Shortcut in RStudio
- Create an R package project in RStudio
- In R console to create a new file: usethis::use_r("set_new_chapter")
- Type and finish the new R function set_new_chapter()
- Create a new folder inst/rstudio. Create a new text file "addins.dcf"
- In R console, type a few R commands to finalize the package and output a package tarball
To use the package in RStudio
- Install the new package as usual
- In RStudio, go to Tools > Modify Keyboard Shortcuts… and search for "dashes". Clicking inside the empty "Shortcut" field and pressing the desired key-combination (eg CMD + d) on your keyboard. Click "Apply". Done
- Go to R editor and type some comments. Now pressing the keyboard shortcut (CMD + d) and dashes will be inserted from the cursor location.
styler
Non-invasive pretty printing of R code https://github.com/r-lib/styler
R> styler:::style_active_file() Using style transformers `styler::tidyverse_style()` The R.cache package needs to create a directory that will hold cache files. It is convenient to use ‘/Users/XXX/Library/Caches/R/R.cache’ because it follows the standard on your operating system and it remains also after restarting R. Do you wish to create the '/Users/XXX/Library/Caches/R/R.cache' directory? If not, a temporary directory (/var/folders/2q/slryb0rx4tj97t66v7l6pwvr_z6g3s/T//RtmplfQTrd/.Rcache) that is specific to this R session will be used. [Y/n]:
remedy: rmarkdown helper
facilitate writing in markdown with RStudio
RegExplain
https://github.com/gadenbuie/regexplain
esquisse
compareWith (meld)
beautifyR
https://github.com/mwip/beautifyR#beautifyr-in-action RStudio addin for formatting Rmarkdown tables and R multi-line comments
Print an R code with line numbers
Use cat -n and the margin option in pandoc.
cat -n mycode.R | pandoc -V geometry:margin=1cm -o mycode.pdf cat -n mycode.R | pandoc -V geometry:"top=1cm, bottom=1.5cm, left=2cm, right=1cm" -o mycode.pdf
Diagnosis
rstudio → run-diagnostics
RStudio Package Manager
- https://docs.rstudio.com/rspm/admin/index.html
- Introduction to the RStudio Package Manager - Sean Lopp
- How do I update packages in RStudio Package Manager?