Statistics: Difference between revisions
Jump to navigation
Jump to search
| Line 41: | Line 41: | ||
Take pomeroy data (7129 x 90) for an example: | Take pomeroy data (7129 x 90) for an example: | ||
<pre> | <pre> | ||
library(gplots) | |||
lr = read.table("C:/ArrayTools/Sample datasets/Pomeroy/Pomeroy -Project/NORMALIZEDLOGINTENSITY.txt") | lr = read.table("C:/ArrayTools/Sample datasets/Pomeroy/Pomeroy -Project/NORMALIZEDLOGINTENSITY.txt") | ||
lr = as.matrix(lr) | lr = as.matrix(lr) | ||
hclust1 <- function(x) hclust(x, method= | method = "average" | ||
hclust1 <- function(x) hclust(x, method= method) | |||
heatmap.2(lr, col=bluered(75), hclustfun = hclust1, distfun = dist, | heatmap.2(lr, col=bluered(75), hclustfun = hclust1, distfun = dist, | ||
density.info="density", scale = "none", | density.info="density", scale = "none", | ||
key=FALSE, symkey=FALSE, trace="none", | key=FALSE, symkey=FALSE, trace="none", | ||
main = | main = method) | ||
</pre> | </pre> | ||
[[File:Hc_ave.png | 250px]] [[File:Hc_com.png | 250px]] [[File:Hc_ward.png | 250px]] | [[File:Hc_ave.png | 250px]] [[File:Hc_com.png | 250px]] [[File:Hc_ward.png | 250px]] | ||
Revision as of 13:59, 1 April 2013
Boxcox transformation
Finding transformation for normal distribution
Visualize the random effects
http://www.quantumforest.com/2012/11/more-sense-of-random-effects/
Sensitivity/Specificity/Accuracy
| Predict | ||||
| 1 | 0 | |||
| True | 1 | TP | FN | Sens=TP/(TP+FN) |
| 0 | FP | TN | Spec=TN/(FP+TN) | |
| N = TP + FP + FN + TN | ||||
- Sensitivity = TP / (TP + FN)
- Specificity = TN / (TN + FP)
- Accuracy = (TP + TN) / N
ROC curve and Brier score
Elements of Statistical Learning
Bagging
Chapter 8 of the book.
- Bootstrap mean is approximately a posterior average.
- Bootstrap aggregation or bagging average: Average the prediction over a collection of bootstrap samples, thereby reducing its variance. The bagging estimate is defined by
- [math]\displaystyle{ \hat{f}_{bag}(x) = \frac{1}{B}\sum_{b=1}^B \hat{f}^{*b}(x). }[/math]
- ksjlfda
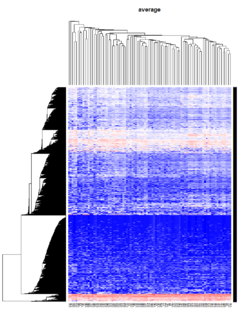
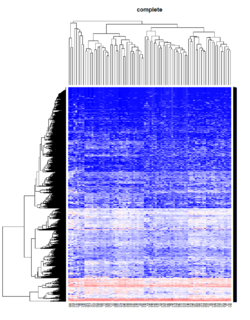
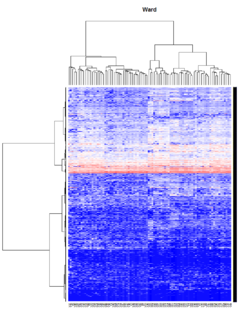
Hierarchical clustering
Take pomeroy data (7129 x 90) for an example:
library(gplots)
lr = read.table("C:/ArrayTools/Sample datasets/Pomeroy/Pomeroy -Project/NORMALIZEDLOGINTENSITY.txt")
lr = as.matrix(lr)
method = "average"
hclust1 <- function(x) hclust(x, method= method)
heatmap.2(lr, col=bluered(75), hclustfun = hclust1, distfun = dist,
density.info="density", scale = "none",
key=FALSE, symkey=FALSE, trace="none",
main = method)