Statistics
Statisticians
- Karl Pearson (1857-1936): chi-square, p-value, PCA
- William Sealy Gosset (1876-1937): Student's t
- Ronald Fisher (1890-1962): ANOVA
- Egon Pearson (1895-1980): son of Karl Pearson
- Jerzy Neyman (1894-1981): type 1 error
Statistics for biologists
http://www.nature.com/collections/qghhqm
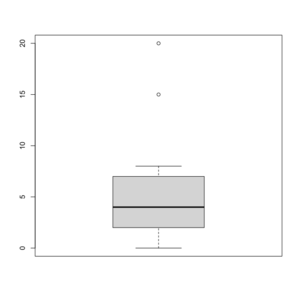
Box/Box and whisker plot in R
See https://flowingdata.com/2008/02/15/how-to-read-and-use-a-box-and-whisker-plot/ for a graphical explanation.
> x=c(0,4,15, 1, 6, 3, 20, 5, 8, 1, 3)
> summary(x)
Min. 1st Qu. Median Mean 3rd Qu. Max.
0 2 4 6 7 20
> sort(x)
[1] 0 1 1 3 3 4 5 6 8 15 20
> boxplot(x, col = 'grey')
- The lower and upper edges of box is determined by the first and 3rd quartiles (2 and 7 in the above example).
- The thick dark horizon line is the median (4 in the example).
- Outliers are defined by observations larger than 3rd quartile + 1.5 * IQR (7+1.5*5=14.5) and smaller than 1st quartile - 1.5 * IQR (2-1.5*5=-5.5). See the empty circles in the plot.
- Upper whisker is defined by the largest data below 3rd quartile + 1.5 * IQR (8 in this example), and the lower whisker is defined by the smallest data greater than 1st quartile - 1.5 * IQR (0 in this example).
Note the wikipedia lists several possible definitions of a whisker. R uses the 2nd method (Tukey boxplot) to define whiskers.
stem and leaf plot
stem(). See R Tutorial.
Note that stem plot is useful when there are outliers.
> stem(x) The decimal point is 10 digit(s) to the right of the | 0 | 00000000000000000000000000000000000000000000000000000000000000000000+419 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 9 > max(x) [1] 129243100275 > max(x)/1e10 [1] 12.92431 > stem(y) The decimal point is at the | 0 | 014478 1 | 0 2 | 1 3 | 9 4 | 8 > y [1] 3.8667356428 0.0001762708 0.7993462430 0.4181079732 0.9541728562 [6] 4.7791262101 0.6899313108 2.1381289177 0.0541736818 0.3868776083 > set.seed(1234) > z <- rnorm(10)*10 > z [1] -12.070657 2.774292 10.844412 -23.456977 4.291247 5.060559 [7] -5.747400 -5.466319 -5.644520 -8.900378 > stem(z) The decimal point is 1 digit(s) to the right of the | -2 | 3 -1 | 2 -0 | 9665 0 | 345 1 | 1
BoxCox transformation
Finding transformation for normal distribution
the Holy Trinity (LRT, Wald, Score tests)
- https://en.wikipedia.org/wiki/Likelihood_function which includes profile likelihood and partial likelihood
- Review of the likelihood theory
- The “Three Plus One” Likelihood-Based Test Statistics: Unified Geometrical and Graphical Interpretations
Don't invert that matrix
- http://www.johndcook.com/blog/2010/01/19/dont-invert-that-matrix/
- http://civilstat.com/2015/07/dont-invert-that-matrix-why-and-how/
Different matrix decompositions/factorizations
- QR decomposition, qr()
- LU decomposition, lu() from the 'Matrix' package
- Cholesky decomposition, chol()
- Singular value decomposition, svd()
set.seed(1234) x <- matrix(rnorm(10*2), nr= 10) cmat <- cov(x); cmat # [,1] [,2] # [1,] 0.9915928 -0.1862983 # [2,] -0.1862983 1.1392095 # cholesky decom d1 <- chol(cmat) t(d1) %*% d1 # equal to cmat d1 # upper triangle # [,1] [,2] # [1,] 0.9957875 -0.1870864 # [2,] 0.0000000 1.0508131 # svd d2 <- svd(cmat) d2$u %*% diag(d2$d) %*% t(d2$v) # equal to cmat d2$u %*% diag(sqrt(d2$d)) # [,1] [,2] # [1,] -0.6322816 0.7692937 # [2,] 0.9305953 0.5226872
Linear Regression
Regression Models for Data Science in R by Brian Caffo
Comic https://xkcd.com/1725/
Different models (in R)
http://www.quantide.com/raccoon-ch-1-introduction-to-linear-models-with-r/
dummy.coef.lm() in R
Extracts coefficients in terms of the original levels of the coefficients rather than the coded variables.
Contrasts in linear regression
- Page 147 of Modern Applied Statistics with S (4th ed)
- https://biologyforfun.wordpress.com/2015/01/13/using-and-interpreting-different-contrasts-in-linear-models-in-r/ This explains the meanings of 'treatment', 'helmert' and 'sum' contrasts.
Multicollinearity
- Multicollinearity in R
- alias: Find Aliases (Dependencies) In A Model
> op <- options(contrasts = c("contr.helmert", "contr.poly"))
> npk.aov <- aov(yield ~ block + N*P*K, npk)
> alias(npk.aov)
Model :
yield ~ block + N * P * K
Complete :
(Intercept) block1 block2 block3 block4 block5 N1 P1 K1 N1:P1 N1:K1 P1:K1
N1:P1:K1 0 1 1/3 1/6 -3/10 -1/5 0 0 0 0 0 0
> options(op)
Confounders
Confidence interval vs prediction interval
Confidence intervals tell you about how well you have determined the mean E(Y). Prediction intervals tell you where you can expect to see the next data point sampled. That is, CI is computed using Var(E(Y|X)) and PI is computed using Var(E(Y|X) + e).
- http://www.graphpad.com/support/faqid/1506/
- http://en.wikipedia.org/wiki/Prediction_interval
- http://robjhyndman.com/hyndsight/intervals/
- https://stat.duke.edu/courses/Fall13/sta101/slides/unit7lec3H.pdf
- https://datascienceplus.com/prediction-interval-the-wider-sister-of-confidence-interval/
Heteroskedasticity
Dealing with heteroskedasticity; regression with robust standard errors using R
Linear regression with Map Reduce
https://freakonometrics.hypotheses.org/53269
Non- and semi-parametric regression
- Cubic and Smoothing Splines in R
- Can we use B-splines to generate non-linear data?
- Semiparametric Regression in R
k-Nearest neighbor regression
- k-NN regression in practice: boundary problem, discontinuities problem.
- Weighted k-NN regression: want weight to be small when distance is large. Common choices - weight = kernel(xi, x)
Kernel regression
- Instead of weighting NN, weight ALL points. Nadaraya-Watson kernel weighted average:
[math]\displaystyle{ \hat{y}_q = \sum c_{qi} y_i/\sum c_{qi} = \frac{\sum \text{Kernel}_\lambda(\text{distance}(x_i, x_q))*y_i}{\sum \text{Kernel}_\lambda(\text{distance}(x_i, x_q))} }[/math].
- Choice of bandwidth [math]\displaystyle{ \lambda }[/math] for bias, variance trade-off. Small [math]\displaystyle{ \lambda }[/math] is over-fitting. Large [math]\displaystyle{ \lambda }[/math] can get an over-smoothed fit. Cross-validation.
- Kernel regression leads to locally constant fit.
- Issues with high dimensions, data scarcity and computational complexity.
Principal component analysis
R source code
> stats:::prcomp.default
function (x, retx = TRUE, center = TRUE, scale. = FALSE, tol = NULL,
...)
{
x <- as.matrix(x)
x <- scale(x, center = center, scale = scale.)
cen <- attr(x, "scaled:center")
sc <- attr(x, "scaled:scale")
if (any(sc == 0))
stop("cannot rescale a constant/zero column to unit variance")
s <- svd(x, nu = 0)
s$d <- s$d/sqrt(max(1, nrow(x) - 1))
if (!is.null(tol)) {
rank <- sum(s$d > (s$d[1L] * tol))
if (rank < ncol(x)) {
s$v <- s$v[, 1L:rank, drop = FALSE]
s$d <- s$d[1L:rank]
}
}
dimnames(s$v) <- list(colnames(x), paste0("PC", seq_len(ncol(s$v))))
r <- list(sdev = s$d, rotation = s$v, center = if (is.null(cen)) FALSE else cen,
scale = if (is.null(sc)) FALSE else sc)
if (retx)
r$x <- x %*% s$v
class(r) <- "prcomp"
r
}
<bytecode: 0x000000003296c7d8>
<environment: namespace:stats>
R example
http://genomicsclass.github.io/book/pages/pca_svd.html
pc <- prcomp(x) group <- as.numeric(tab$Tissue) plot(pc$x[, 1], pc$x[, 2], col = group, main = "PCA", xlab = "PC1", ylab = "PC2")
The meaning of colors can be found by palette().
- black
- red
- green3
- blue
- cyan
- magenta
- yellow
- gray
PCA and SVD
Using the SVD to perform PCA makes much better sense numerically than forming the covariance matrix to begin with, since the formation of XX⊤ can cause loss of precision.
http://math.stackexchange.com/questions/3869/what-is-the-intuitive-relationship-between-svd-and-pca
AIC/BIC in estimating the number of components
Related to Factor Analysis
- http://www.aaronschlegel.com/factor-analysis-introduction-principal-component-method-r/.
- http://support.minitab.com/en-us/minitab/17/topic-library/modeling-statistics/multivariate/principal-components-and-factor-analysis/differences-between-pca-and-factor-analysis/
In short,
- In Principal Components Analysis, the components are calculated as linear combinations of the original variables. In Factor Analysis, the original variables are defined as linear combinations of the factors.
- In Principal Components Analysis, the goal is to explain as much of the total variance in the variables as possible. The goal in Factor Analysis is to explain the covariances or correlations between the variables.
- Use Principal Components Analysis to reduce the data into a smaller number of components. Use Factor Analysis to understand what constructs underlie the data.
Calculated by Hand
http://strata.uga.edu/software/pdf/pcaTutorial.pdf
Do not scale your matrix
https://privefl.github.io/blog/(Linear-Algebra)-Do-not-scale-your-matrix/
Visualization
- PCA and Visualization
- Scree plots from the FactoMineR package (based on ggplot2)
What does it do if we choose center=FALSE in prcomp()?
In USArrests data, use center=FALSE gives a better scatter plot of the first 2 PCA components.
x1 = prcomp(USArrests) x2 = prcomp(USArrests, center=F) plot(x1$x[,1], x1$x[,2]) # looks random windows(); plot(x2$x[,1], x2$x[,2]) # looks good in some sense
Relation to Multidimensional scaling/MDS
With no missing data, classical MDS (Euclidean distance metric) is the same as PCA.
Comparisons are here.
Differences are asked/answered on stackexchange.com. The post also answered the question when these two are the same.
Matrix factorization methods
http://joelcadwell.blogspot.com/2015/08/matrix-factorization-comes-in-many.html Review of principal component analysis (PCA), K-means clustering, nonnegative matrix factorization (NMF) and archetypal analysis (AA).
Partial Least Squares (PLS)
- https://en.wikipedia.org/wiki/Partial_least_squares_regression
- Supervised vs. Unsupervised Learning: Exploring Brexit with PLS and PCA
- pls R package
- plsRcox R package (archived). See here for the installation.
PLS, PCR (principal components regression) and ridge regression tend to behave similarly. Ridge regression may be preferred because it shrinks smoothly, rather than in discrete steps.
Independent component analysis
ICA is another dimensionality reduction method.
ICA vs PCA
ICS vs FA
Correspondence analysis
https://francoishusson.wordpress.com/2017/07/18/multiple-correspondence-analysis-with-factominer/ and the book Exploratory Multivariate Analysis by Example Using R
t-SNE
t-Distributed Stochastic Neighbor Embedding (t-SNE) is a technique for dimensionality reduction that is particularly well suited for the visualization of high-dimensional datasets.
- https://distill.pub/2016/misread-tsne/
- https://lvdmaaten.github.io/tsne/
- Application to ARCHS4
- Visualization of High Dimensional Data using t-SNE with R
Visualize the random effects
http://www.quantumforest.com/2012/11/more-sense-of-random-effects/
ROC curve and Brier score
- Binary case:
- Y = true positive rate = sensitivity,
- X = false positive rate = 1-specificity
- Calibration
- Calculate AUC by hand. AUC is equal to the probability that a true positive is scored greater than a true negative. See the proof on wikipedia.
- How to calculate Area Under the Curve (AUC), or the c-statistic, by hand or by R
- Introduction to the ROCR package.
- http://freakonometrics.hypotheses.org/9066, http://freakonometrics.hypotheses.org/20002
- Illustrated Guide to ROC and AUC
- ROC Curves in Two Lines of R Code
- Gini and AUC. Gini = 2*AUC-1.
- 'Survival Model Predictive Accuracy and ROC Curves' by Heagerty & Zheng 2005
- Sensitivity [math]\displaystyle{ P(\hat{p_i} \gt c | Y_i=1) }[/math], Specificity [math]\displaystyle{ P(\hat{p}_i \le c | Y_i=0 }[/math]), [math]\displaystyle{ Y_i }[/math] is binary outcomes, [math]\displaystyle{ \hat{p}_i }[/math] is a prediction, [math]\displaystyle{ c }[/math] is a criterion for classifying the prediction as positive ([math]\displaystyle{ \hat{p}_i \gt c }[/math]) or negative ([math]\displaystyle{ \hat{p}_i \le c }[/math]).
- The AUC measures the probability that the marker value for a randomly selected case exceeds the marker value for a randomly selected control
- ROC curves are useful for comparing the discriminatory capacity of different potential biomarkers.
Confusion matrix, Sensitivity/Specificity/Accuracy
| Predict | ||||
| 1 | 0 | |||
| True | 1 | TP | FN | Sens=TP/(TP+FN) FNR=FN/(TP+FN) |
| 0 | FP | TN | Spec=TN/(FP+TN) | |
| PPV=TP/(TP+FP) FDR=FP/(TP+FP) |
NPV=TN/(FN+TN) | N = TP + FP + FN + TN | ||
- Sensitivity = TP / (TP + FN)
- Specificity = TN / (TN + FP)
- Accuracy = (TP + TN) / N
- False discovery rate FDR = FP / (TP + FP)
- False negative rate FNR = FN / (TP + FN)
- Positive predictive value (PPV) = TP / # positive calls = TP / (TP + FP) = 1 - FDR
- Negative predictive value (NPV) = TN / # negative calls = TN / (FN + TN)
- Prevalence = (TP + FN) / N.
- Note that PPV & NPV can also be computed from sensitivity, specificity, and prevalence:
- PPV is directly proportional to the prevalence of the disease or condition..
- For example, in the extreme case if the prevalence =1, then PPV is always 1.
- [math]\displaystyle{ \text{PPV} = \frac{\text{sensitivity} \times \text{prevalence}}{\text{sensitivity} \times \text{prevalence}+(1-\text{specificity}) \times (1-\text{prevalence})} }[/math]
- [math]\displaystyle{ \text{NPV} = \frac{\text{specificity} \times (1-\text{prevalence})}{(1-\text{sensitivity}) \times \text{prevalence}+\text{specificity} \times (1-\text{prevalence})} }[/math]
Precision recall curve
- Precision and recall
- Y axis = Precision = tp/(tp + fp) = PPV, large is better
- X axis = Recall = tp/(tp + fn) = Sensitivity, large is better
- The Relationship Between Precision-Recall and ROC Curves
Incidence, Prevalence
https://www.health.ny.gov/diseases/chronic/basicstat.htm
genefilter package and rowpAUCs function
- rowpAUCs function in genefilter package. The aim is to find potential biomarkers whose expression level is able to distinguish between two groups.
# source("http://www.bioconductor.org/biocLite.R")
# biocLite("genefilter")
library(Biobase) # sample.ExpressionSet data
data(sample.ExpressionSet)
library(genefilter)
r2 = rowpAUCs(sample.ExpressionSet, "sex", p=0.1)
plot(r2[1]) # first gene, asking specificity = .9
r2 = rowpAUCs(sample.ExpressionSet, "sex", p=1.0)
plot(r2[1]) # it won't show pAUC
r2 = rowpAUCs(sample.ExpressionSet, "sex", p=.999)
plot(r2[1]) # pAUC is very close to AUC now
Use and Misuse of the Receiver Operating Characteristic Curve in Risk Prediction
http://circ.ahajournals.org/content/115/7/928
Performance evaluation
- Testing for improvement in prediction model performance by Pepe et al 2013.
Maximum likelihood
Difference of partial likelihood, profile likelihood and marginal likelihood
Generalized Linear Model
Lectures from a course in Simon Fraser University Statistics.
Doing magic and analyzing seasonal time series with GAM (Generalized Additive Model) in R
Quasi Likelihood
Quasi-likelihood is like log-likelihood. The quasi-score function (first derivative of quasi-likelihood function) is the estimating equation.
- Original paper by Peter McCullagh.
- Lecture 20 from SFU.
- U. Washington and another lecture focuses on overdispersion.
- This lecture contains a table of quasi likelihood from common distributions.
Plot
Deviance, stats::deviance() and glmnet::deviance.glmnet() from R
- It is a generalization of the idea of using the sum of squares of residuals (RSS) in ordinary least squares to cases where model-fitting is achieved by maximum likelihood. See What is Deviance? (specifically in CART/rpart) to manually compute deviance and compare it with the returned value of the deviance() function from a linear regression. Summary: deviance() = RSS in linear models.
- https://www.rdocumentation.org/packages/stats/versions/3.4.3/topics/deviance
- Likelihood ratio tests and the deviance http://data.princeton.edu/wws509/notes/a2.pdf#page=6
- Deviance(y,muhat) = 2*(loglik_saturated - loglik_proposed)
- Interpreting Residual and Null Deviance in GLM R
- Null Deviance = 2(LL(Saturated Model) - LL(Null Model)) on df = df_Sat - df_Null. The null deviance shows how well the response variable is predicted by a model that includes only the intercept (grand mean).
- Residual Deviance = 2(LL(Saturated Model) - LL(Proposed Model)) = [math]\displaystyle{ 2(LL(y|y) - LL(\hat{\mu}|y)) }[/math], df = df_Sat - df_Proposed=n-p. ==> deviance() has returned.
- Null deviance > Residual deviance. Null deviance df = n-1. Residual deviance df = n-p.
## an example with offsets from Venables & Ripley (2002, p.189)
utils::data(anorexia, package = "MASS")
anorex.1 <- glm(Postwt ~ Prewt + Treat + offset(Prewt),
family = gaussian, data = anorexia)
summary(anorex.1)
# Call:
# glm(formula = Postwt ~ Prewt + Treat + offset(Prewt), family = gaussian,
# data = anorexia)
#
# Deviance Residuals:
# Min 1Q Median 3Q Max
# -14.1083 -4.2773 -0.5484 5.4838 15.2922
#
# Coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 49.7711 13.3910 3.717 0.000410 ***
# Prewt -0.5655 0.1612 -3.509 0.000803 ***
# TreatCont -4.0971 1.8935 -2.164 0.033999 *
# TreatFT 4.5631 2.1333 2.139 0.036035 *
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#
# (Dispersion parameter for gaussian family taken to be 48.69504)
#
# Null deviance: 4525.4 on 71 degrees of freedom
# Residual deviance: 3311.3 on 68 degrees of freedom
# AIC: 489.97
#
# Number of Fisher Scoring iterations: 2
deviance(anorex.1)
# [1] 3311.263
- In glmnet package. The deviance is defined to be 2*(loglike_sat - loglike), where loglike_sat is the log-likelihood for the saturated model (a model with a free parameter per observation). Null deviance is defined to be 2*(loglike_sat -loglike(Null)); The NULL model refers to the intercept model, except for the Cox, where it is the 0 model. Hence dev.ratio=1-deviance/nulldev, and this deviance method returns (1-dev.ratio)*nulldev.
x=matrix(rnorm(100*2),100,2) y=rnorm(100) fit1=glmnet(x,y) deviance(fit1) # one for each lambda # [1] 98.83277 98.53893 98.29499 98.09246 97.92432 97.78472 97.66883 # [8] 97.57261 97.49273 97.41327 97.29855 97.20332 97.12425 97.05861 # ... # [57] 96.73772 96.73770 fit2 <- glmnet(x, y, lambda=.1) # fix lambda deviance(fit2) # [1] 98.10212 deviance(glm(y ~ x)) # [1] 96.73762 sum(residuals(glm(y ~ x))^2) # [1] 96.73762
Saturated model
- The saturated model always has n parameters where n is the sample size.
- Logistic Regression : How to obtain a saturated model
Simulate data
Density plot
# plot a Weibull distribution with shape and scale func <- function(x) dweibull(x, shape = 1, scale = 3.38) curve(func, .1, 10) func <- function(x) dweibull(x, shape = 1.1, scale = 3.38) curve(func, .1, 10)
The shape parameter plays a role on the shape of the density function and the failure rate.
- Shape <=1: density is convex, not a hat shape.
- Shape =1: failure rate (hazard function) is constant. Exponential distribution.
- Shape >1: failure rate increases with time
Simulate data from a specified density
Signal to noise ratio
- https://en.wikipedia.org/wiki/Signal-to-noise_ratio
- https://stats.stackexchange.com/questions/31158/how-to-simulate-signal-noise-ratio
- [math]\displaystyle{ \frac{\sigma^2_{signal}}{\sigma^2_{noise}} = \frac{Var(f(X))}{Var(e)} }[/math] if Y = f(X) + e
- Page 401 of ESLII (https://web.stanford.edu/~hastie/ElemStatLearn//) 12th print.
Some examples of signal to noise ratio
- ESLII_print12.pdf: .64, 5, 4
- Yuan and Lin 2006: 1.8, 3
Effect size
- [math]\displaystyle{ \theta = \frac{\mu_1 - \mu_2} \sigma, }[/math]
See also the estimation by the pooled sd.
Multiple comparisons
- If you perform experiments over and over, you's bound to find something. So significance level must be adjusted down when performing multiple hypothesis tests.
- http://www.gs.washington.edu/academics/courses/akey/56008/lecture/lecture10.pdf
- Book 'Multiple Comparison Using R' by Bretz, Hothorn and Westfall, 2011.
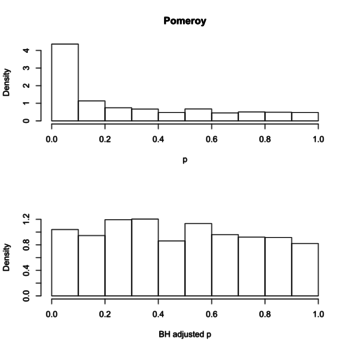
- Plot a histogram of p-values, a post from varianceexplained.org. The anti-conservative histogram (tail on the RHS) is what we have typically seen in e.g. microarray gene expression data.
- Comparison of different ways of multiple-comparison in R.
Take an example, Suppose 550 out of 10,000 genes are significant at .05 level
- P-value < .05 ==> Expect .05*10,000=500 false positives
- False discovery rate < .05 ==> Expect .05*550 =27.5 false positives
- Family wise error rate < .05 ==> The probablity of at least 1 false positive <.05
According to Lifetime Risk of Developing or Dying From Cancer, there is a 39.7% risk of developing a cancer for male during his lifetime (in other words, 1 out of every 2.52 men in US will develop some kind of cancer during his lifetime) and 37.6% for female. So the probability of getting at least one cancer patient in a 3-generation family is 1-.6**3 - .63**3 = 0.95.
False Discovery Rate
- https://en.wikipedia.org/wiki/False_discovery_rate
- Paper Definition by Benjamini and Hochberg in JRSS B 1995.
- A comic
- Statistical significance for genomewide studies by Storey and Tibshirani.
- What’s the probability that a significant p-value indicates a true effect?
- http://onetipperday.sterding.com/2015/12/my-note-on-multiple-testing.html
Suppose [math]\displaystyle{ p_1 \leq p_2 \leq ... \leq p_n }[/math]. Then
- [math]\displaystyle{ \text{FDR}_i = \text{min}(1, n* p_i/i) }[/math].
So if the number of tests ([math]\displaystyle{ n }[/math]) is large and/or the original p value ([math]\displaystyle{ p_i }[/math]) is large, then FDR can hit the value 1.
However, the simple formula above does not guarantee the monotonicity property from the FDR. So the calculation in R is more complicated. See How Does R Calculate the False Discovery Rate.
Below is the histograms of p-values and FDR (BH adjusted) from a real data (Pomeroy in BRB-ArrayTools).
q-value
q-value is defined as the minimum FDR that can be attained when calling that feature significant (i.e., expected proportion of false positives incurred when calling that feature significant).
If gene X has a q-value of 0.013 it means that 1.3% of genes that show p-values at least as small as gene X are false positives.
SAM/Significance Analysis of Microarrays
The percentile option is used to define the number of falsely called genes based on 'B' permutations. If we use the 90-th percentile, the number of significant genes will be less than if we use the 50-th percentile/median.
In BRCA dataset, using the 90-th percentile will get 29 genes vs 183 genes if we use median.
Multivariate permutation test
In BRCA dataset, using 80% confidence gives 116 genes vs 237 genes if we use 50% confidence (assuming maximum proportion of false discoveries is 10%). The method is published on EL Korn, JF Troendle, LM McShane and R Simon, Controlling the number of false discoveries: Application to high dimensional genomic data, Journal of Statistical Planning and Inference, vol 124, 379-398 (2004).
String Permutations Algorithm
Bayes
Bayes factor
Empirical Bayes method
Naive Bayes classifier
Understanding Naïve Bayes Classifier Using R
MCMC
Speeding up Metropolis-Hastings with Rcpp
Offset in Poisson regression
https://stats.stackexchange.com/questions/11182/when-to-use-an-offset-in-a-poisson-regression
- We need to model rates instead of counts
- More generally, you use offsets because the units of observation are different in some dimension (different populations, different geographic sizes) and the outcome is proportional to that dimension.
offset() function
- An offset is a term to be added to a linear predictor, such as in a generalised linear model, with known coefficient 1 rather than an estimated coefficient.
- https://www.rdocumentation.org/packages/stats/versions/3.5.0/topics/offset
- An example from here
Y <- c(15, 7, 36, 4, 16, 12, 41, 15) N <- c(4949, 3534, 12210, 344, 6178, 4883, 11256, 7125) x1 <- c(-0.1, 0, 0.2, 0, 1, 1.1, 1.1, 1) x2 <- c(2.2, 1.5, 4.5, 7.2, 4.5, 3.2, 9.1, 5.2) glm(Y ~ offset(log(N)) + (x1 + x2), family=poisson) # two variables # Coefficients: # (Intercept) x1 x2 # -6.172 -0.380 0.109 # # Degrees of Freedom: 7 Total (i.e. Null); 5 Residual # Null Deviance: 10.56 # Residual Deviance: 4.559 AIC: 46.69 glm(Y ~ offset(log(N)) + I(x1+x2), family=poisson) # one variable # Coefficients: # (Intercept) I(x1 + x2) # -6.12652 0.04746 # # Degrees of Freedom: 7 Total (i.e. Null); 6 Residual # Null Deviance: 10.56 # Residual Deviance: 8.001 AIC: 48.13
Overdispersion
https://en.wikipedia.org/wiki/Overdispersion
Var(Y) = phi * E(Y). If phi > 1, then it is overdispersion relative to Poisson. If phi <1, we have under-dispersion (rare).
Heterogeneity
The Poisson model fit is not good; residual deviance/df >> 1. The lack of fit maybe due to missing data, covariates or overdispersion.
Subjects within each covariate combination still differ greatly.
- https://onlinecourses.science.psu.edu/stat504/node/169.
- https://onlinecourses.science.psu.edu/stat504/node/162
Consider Quasi-Poisson or negative binomial.
Test of overdispersion or underdispersion in Poisson models
Negative Binomial
The mean of the Poisson distribution can itself be thought of as a random variable drawn from the gamma distribution thereby introducing an additional free parameter.
Zero counts
Survival data
- https://web.stanford.edu/~lutian/coursepdf/stat331.HTML and https://web.stanford.edu/~lutian/coursepdf/ (3 types of tests).
- http://www.stat.columbia.edu/~madigan/W2025/notes/survival.pdf.
- How to manually compute the KM curve and by R
- Estimation of parametric survival function from joint likelihood in theory and R.
- http://data.princeton.edu/wws509/notes/c7s1.html
- http://data.princeton.edu/pop509/ParametricSurvival.pdf Parametric survival models with covariates (logT = alpha + sigma W) p8
- Weibull p2 where T ~ Weibull and W ~ Extreme value.
- Gamma p3 where T ~ Gamma and W ~ Generalized extreme value
- Generalized gamma p4,
- log normal p4 where T ~ lognormal and W ~ N(0,1)
- log logistic p4 where T ~ log logistic and W ~ standard logistic distribution.
- http://www.math.ucsd.edu/~rxu/math284/ (good cover) Wald test
- http://www.stats.ox.ac.uk/~mlunn/
- https://www.openintro.org/download.php?file=survival_analysis_in_R&referrer=/stat/surv.php
- https://cran.r-project.org/web/packages/survival/vignettes/timedep.pdf
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1065034/
- Survival Analysis with R from rviews.rstudio.com
- Survival Analysis with R from bioconnector.og.
Censoring
http://sphweb.bumc.bu.edu/otlt/MPH-Modules/BS/BS704_Survival/BS704_Survival_print.html
The most common is called right censoring and occurs when a participant does not have the event of interest during the study and thus their last observed follow-up time is less than their time to event. This can occur when a participant drops out before the study ends or when a participant is event free at the end of the observation period.
Definitions of common terms in survival analysis
- Event: Death, disease occurrence, disease recurrence, recovery, or other experience of interest
- Time: The time from the beginning of an observation period (such as surgery or beginning treatment) to (i) an event, or (ii) end of the study, or (iii) loss of contact or withdrawal from the study.
- Censoring / Censored observation: If a subject does not have an event during the observation time, they are described as censored. The subject is censored in the sense that nothing is observed or known about that subject after the time of censoring. A censored subject may or may not have an event after the end of observation time.
In R, "status" should be called "event status". status = 1 means event occurred. status =0 means no event (censored).
How to explore survival data
https://en.wikipedia.org/wiki/Survival_analysis#Survival_analysis_in_R
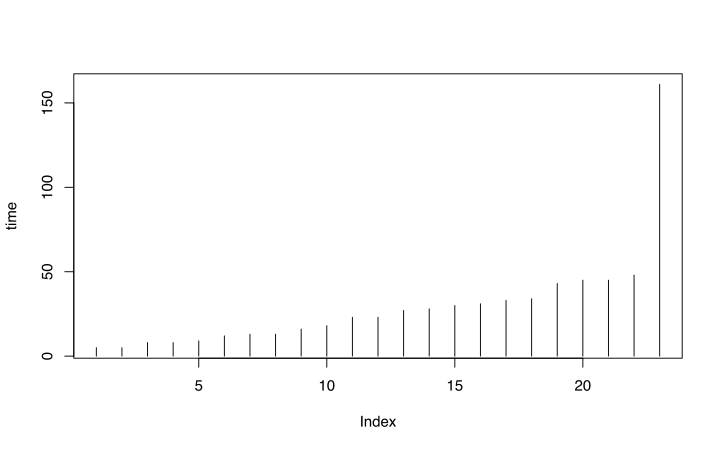
- Create graph of length of time that each subject was in the study
library(survival) # sort the aml data by time aml <- aml[order(aml$time),] with(aml, plot(time, type="h"))
- Create the life table survival object
summary(aml.survfit)
Call: survfit(formula = Surv(time, status == 1) ~ 1, data = aml)
time n.risk n.event survival std.err lower 95% CI upper 95% CI
5 23 2 0.9130 0.0588 0.8049 1.000
8 21 2 0.8261 0.0790 0.6848 0.996
9 19 1 0.7826 0.0860 0.6310 0.971
12 18 1 0.7391 0.0916 0.5798 0.942
13 17 1 0.6957 0.0959 0.5309 0.912
18 14 1 0.6460 0.1011 0.4753 0.878
23 13 2 0.5466 0.1073 0.3721 0.803
27 11 1 0.4969 0.1084 0.3240 0.762
30 9 1 0.4417 0.1095 0.2717 0.718
31 8 1 0.3865 0.1089 0.2225 0.671
33 7 1 0.3313 0.1064 0.1765 0.622
34 6 1 0.2761 0.1020 0.1338 0.569
43 5 1 0.2208 0.0954 0.0947 0.515
45 4 1 0.1656 0.0860 0.0598 0.458
48 2 1 0.0828 0.0727 0.0148 0.462
- Kaplan-Meier curve for aml with the confidence bounds.
plot(aml.survfit, xlab = "Time", ylab="Proportion surviving")
- Create aml life tables broken out by treatment (x, "Maintained" vs. "Not maintained")
surv.by.aml.rx <- survfit(Surv(time, status == 1) ~ x, data = aml)
summary(surv.by.aml.rx)
Call: survfit(formula = Surv(time, status == 1) ~ x, data = aml)
x=Maintained
time n.risk n.event survival std.err lower 95% CI upper 95% CI
9 11 1 0.909 0.0867 0.7541 1.000
13 10 1 0.818 0.1163 0.6192 1.000
18 8 1 0.716 0.1397 0.4884 1.000
23 7 1 0.614 0.1526 0.3769 0.999
31 5 1 0.491 0.1642 0.2549 0.946
34 4 1 0.368 0.1627 0.1549 0.875
48 2 1 0.184 0.1535 0.0359 0.944
x=Nonmaintained
time n.risk n.event survival std.err lower 95% CI upper 95% CI
5 12 2 0.8333 0.1076 0.6470 1.000
8 10 2 0.6667 0.1361 0.4468 0.995
12 8 1 0.5833 0.1423 0.3616 0.941
23 6 1 0.4861 0.1481 0.2675 0.883
27 5 1 0.3889 0.1470 0.1854 0.816
30 4 1 0.2917 0.1387 0.1148 0.741
33 3 1 0.1944 0.1219 0.0569 0.664
43 2 1 0.0972 0.0919 0.0153 0.620
45 1 1 0.0000 NaN NA NA
- Plot KM plot broken out by treatment
plot(surv.by.aml.rx, xlab = "Time", ylab="Survival",
col=c("black", "red"), lty = 1:2,
main="Kaplan-Meier Survival vs. Maintenance in AML")
legend(100, .6, c("Maintained", "Not maintained"),
lty = 1:2, col=c("black", "red"))
- Perform the log rank test using the R function survdiff().
surv.diff.aml <- survdiff(Surv(time, status == 1) ~ x, data=aml)
surv.diff.aml
Call:
survdiff(formula = Surv(time, status == 1) ~ x, data = aml)
N Observed Expected (O-E)^2/E (O-E)^2/V
x=Maintained 11 7 10.69 1.27 3.4
x=Nonmaintained 12 11 7.31 1.86 3.4
Chisq= 3.4 on 1 degrees of freedom, p= 0.07
Kaplan & Meier and Nelson-Aalen: survfit.formula()
- Landmarks
- Kaplan-Meier: 1958
- Nelson: 1969
- Cox and Brewlow: 1972 S(t) = exp(-Lambda(t))
- Aalen: 1978 Lambda(t)
- D distinct times [math]\displaystyle{ t_1 \lt t_2 \lt \cdots \lt t_D }[/math]. At time [math]\displaystyle{ t_i }[/math] there are [math]\displaystyle{ d_i }[/math] events. Let [math]\displaystyle{ Y_i }[/math] be the number of individuals who are at risk at time [math]\displaystyle{ t_i }[/math]. The quantity [math]\displaystyle{ d_i/Y_i }[/math] provides an estimate of the conditional probability that an individual who survives to just prior to time [math]\displaystyle{ t_i }[/math] experiences the event at time [math]\displaystyle{ t_i }[/math]. The KM estimator of the survival function and the Nelson-Aalen estimator of the cumulative hazard are define as follows ([math]\displaystyle{ t_1 \le t }[/math]):
- [math]\displaystyle{ \begin{align} \hat{S}(t) &= \prod_{t_i \le t} [1 - d_i/Y_i] \\ \hat{H}(t) &= \sum_{t_i \le t} d_i/Y_i \end{align} }[/math]
str(kidney)
'data.frame': 76 obs. of 7 variables:
$ id : num 1 1 2 2 3 3 4 4 5 5 ...
$ time : num 8 16 23 13 22 28 447 318 30 12 ...
$ status : num 1 1 1 0 1 1 1 1 1 1 ...
$ age : num 28 28 48 48 32 32 31 32 10 10 ...
$ sex : num 1 1 2 2 1 1 2 2 1 1 ...
$ disease: Factor w/ 4 levels "Other","GN","AN",..: 1 1 2 2 1 1 1 1 1 1 ...
$ frail : num 2.3 2.3 1.9 1.9 1.2 1.2 0.5 0.5 1.5 1.5 ...
kidney[order(kidney$time), c("time", "status")]
kidney[kidney$time == 13, ] # one is dead and the other is alive
length(unique(kidney$time)) # 60
sfit <- survfit(Surv(time, status) ~ 1, data = kidney)
str(sfit)
List of 13
$ n : int 76
$ time : num [1:60] 2 4 5 6 7 8 9 12 13 15 ...
$ n.risk : num [1:60] 76 75 74 72 71 69 65 64 62 60 ...
$ n.event : num [1:60] 1 0 0 0 2 2 1 2 1 2 ...
$ n.censor : num [1:60] 0 1 2 1 0 2 0 0 1 0 ...
$ surv : num [1:60] 0.987 0.987 0.987 0.987 0.959 ...
$ type : chr "right"
all(sapply(sfit$time, function(tt) sum(kidney$time >= tt)) == sfit$n.risk) # TRUE
all(sapply(sfit$time, function(tt) sum(kidney$status[kidney$time == tt])) == sfit$n.event) # TRUE
all(sapply(sfit$time, function(tt) sum(1-kidney$status[kidney$time == tt])) == sfit$n.censor) # TRUE
all(cumprod(1 - sfit$n.event/sfit$n.risk) == sfit$surv) # FALSE
range(abs(cumprod(1 - sfit$n.event/sfit$n.risk) - sfit$surv))
# [1] 0.000000e+00 1.387779e-17
- Note that the KM estimate is left-continuous step function with the intervals closed at left and open at right. For [math]\displaystyle{ t \in [t_j, t_{j+1}) }[/math] for a certain j, we have [math]\displaystyle{ \hat{S}(t) = \prod_{i=1}^j (1-d_i/n_i) }[/math] where [math]\displaystyle{ d_i }[/math] is the number people who have an event during the interval [math]\displaystyle{ [t_i, t_{i+1}) }[/math] and [math]\displaystyle{ n_i }[/math] is the number of people at risk just before the beginning of the interval [math]\displaystyle{ [t_i, t_{i+1}) }[/math].
- The product-limit estimator can be constructed by using a reduced-sample approach. We can estimate the [math]\displaystyle{ P(T \gt t_i | T \ge t_i) = \frac{Y_i - d_i}{Y_i} }[/math] for [math]\displaystyle{ i=1,2,\cdots,D }[/math]. [math]\displaystyle{ S(t_i) = \frac{S(t_i)}{S(t_{i-1})} \frac{S(t_{i-1})}{S(t_{i-2})} \cdots \frac{S(t_2)}{S(t_1)} \frac{S(t_1)}{S(0)} S(0) = P(T \gt t_i | T \ge t_i) P(T \gt t_{i-1} | T \ge t_{i-1}) \cdots P(T\gt t_2|T \ge t_2) P(T\gt t_1 | T \ge t_1) }[/math] because S(0)=1 and, for a discrete distribution, [math]\displaystyle{ S(t_{i-1}) = P(T \gt t_{i-1}) = P(T \ge t_i) }[/math].
- Self consistency. If we had no censored observations, the estimator of the survival function at a time t is the proportion of observations which are larger than t, that is, [math]\displaystyle{ \hat{S}(t) = \frac{1}{n}\sum I(X_i \gt t) }[/math].
- Curves are plotted in the same order as they are listed by print (which gives a 1 line summary of each). For example, -1 < 1 and 'Maintenance' < 'Nonmaintained'. That means, the labels list in the legend() command should have the same order as the curves.
- Kaplan and Meier is used to give an estimator of the survival function S(t)
- Nelson-Aalen estimator is for the cumulative hazard H(t). Note that [math]\displaystyle{ 0 \le H(t) \lt \infty }[/math] and [math]\displaystyle{ H(t) \rightarrow \infty }[/math] as t goes to infinity. So there is a constraint on the hazard function, see Wikipedia.
Note that S(t) is related to H(t) by [math]\displaystyle{ H(t) = -ln[S(t)]. }[/math] The two estimators are similar (see example 4.1A and 4.1B from Klein and Moeschberge).
The Nelson-Aalen estimator has two primary uses in analyzing data
- Selecting between parametric models for the time to event
- Crude estimates of the hazard rate h(t). This is related to the estimation of the survival function in Cox model. See 8.6 of Klein and Moeschberge.
The Kaplan–Meier estimator (the product limit estimator) is an estimator for estimating the survival function from lifetime data. In medical research, it is often used to measure the fraction of patients living for a certain amount of time after treatment.
The "+" sign means censored observations and a long vertical line (not '+') means there is a dead observation at that time.
If the last observation (longest survival time) is dead, the survival curve will goes down to zero. Otherwise, the survival curve will remain flat.
Usually the KM curve of treatment group is higher than that of the control group.
The Y-axis (the probability that a member from a given population will have a lifetime exceeding time) is often called
- Cumulative probability
- Cumulative survival
- Percent survival
- Probability without event
- Proportion alive/surviving
- Survival
- Survival probability
> library(survival)
> str(aml$x)
Factor w/ 2 levels "Maintained","Nonmaintained": 1 1 1 1 1 1 1 1 1 1 ...
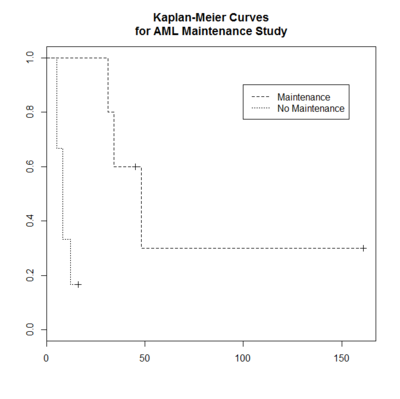
> plot(leukemia.surv <- survfit(Surv(time, status) ~ x, data = aml[7:17,] ) ,
lty=2:3, mark.time = TRUE) # a (small) subset, mark.time is used to show censored obs
> aml[7:17,]
time status x
7 31 1 Maintained
8 34 1 Maintained
9 45 0 Maintained
10 48 1 Maintained
11 161 0 Maintained
12 5 1 Nonmaintained
13 5 1 Nonmaintained
14 8 1 Nonmaintained
15 8 1 Nonmaintained
16 12 1 Nonmaintained
17 16 0 Nonmaintained
> legend(100, .9, c("Maintenance", "No Maintenance"), lty = 2:3) # lty: 2=dashed, 3=dotted
> title("Kaplan-Meier Curves\nfor AML Maintenance Study")
# Cumulative hazard plot
# Lambda(t) = -log(S(t));
# see https://en.wikipedia.org/wiki/Survival_analysis
# http://statweb.stanford.edu/~olshen/hrp262spring01/spring01Handouts/Phil_doc.pdf
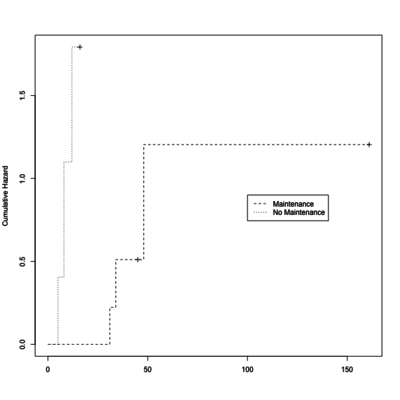
plot(leukemia.surv <- survfit(Surv(time, status) ~ x, data = aml[7:17,] ) ,
lty=2:3, mark.time = T, fun="cumhaz", ylab="Cumulative Hazard")
- Kaplan-Meier estimator from the wikipedia.
- Two papers this and this to describe steps to calculate the KM estimate.
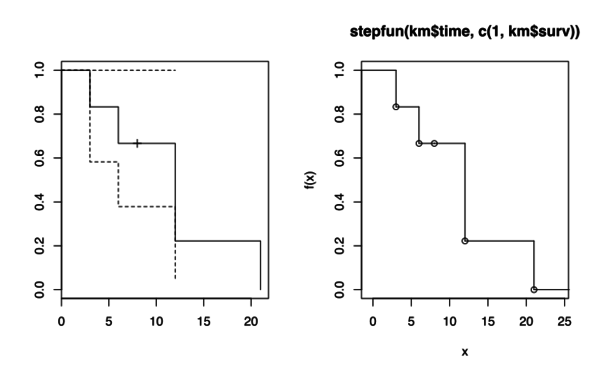
- Estimating a survival probability in R
# https://www.lexjansen.com/pharmasug/2011/CC/PharmaSUG-2011-CC16.pdf mydata <- data.frame(time=c(3,6,8,12,12,21),status=c(1,1,0,1,1,1)) km <- survfit(Surv(time, status)~1, data=mydata) plot(km, mark.time = T) survest <- stepfun(km$time, c(1, km$surv)) plot(survest) > str(km) List of 13 $ n : int 6 $ time : num [1:5] 3 6 8 12 21 $ n.risk : num [1:5] 6 5 4 3 1 $ n.event : num [1:5] 1 1 0 2 1 $ n.censor : num [1:5] 0 0 1 0 0 $ surv : num [1:5] 0.833 0.667 0.667 0.222 0 $ type : chr "right" $ std.err : num [1:5] 0.183 0.289 0.289 0.866 Inf $ upper : num [1:5] 1 1 1 1 NA $ lower : num [1:5] 0.5827 0.3786 0.3786 0.0407 NA $ conf.type: chr "log" $ conf.int : num 0.95
Multiple curves
Curves/groups are ordered. The first color in the palette is used to color the first level of the factor variable. This is same idea as ggsurvplot in the survminer package. This affects parameters like col and lty in plot() function. For example,
- 1<2
- 'c' < 't'
- 'control' < 'treatment'
- 'Control' < 'Treatment'
- 'female' < 'male'.
For legend(), the first category in legend argument will appear at the top of the legend box.
Inverse Probability of Censoring Weighted (IPCW)
- Inverse probability weighting from Wikipedia
- https://www.bmj.com/content/352/bmj.i189.full.print Four examples are considered.
- Correcting for Noncompliance and Dependent Censoring in an AIDS Clinical Trial with Inverse Probability of Censoring Weighted (IPCW) Log‐Rank Tests by James M. Robins, Biometrics 2000.
- The Kaplan–Meier Estimator as an Inverse-Probability-of-Censoring Weighted Average by Satten 2001. IPCW.
The plots below show by flipping the status variable, we can recover the survival function of the censoring variable.
require(survival)
n = 10000
beta1 = 2; beta2 = -1
lambdaT = 1 # baseline hazard
lambdaC = 2 # hazard of censoring
set.seed(1234)
x1 = rnorm(n,0)
x2 = rnorm(n,0)
# true event time
T = rweibull(n, shape=1, scale=lambdaT*exp(-beta1*x1-beta2*x2))
# method 1: exponential censoring variable
C <- rweibull(n, shape=1, scale=lambdaC)
time = pmin(T,C)
status <- 1*(T <= C)
mean(status)
summary(T)
summary(C)
par(mfrow=c(2,1), mar = c(3,4,2,2)+.1)
status2 <- 1-status
plot(survfit(Surv(time, status2) ~ 1),
ylab="Survival probability",
main = 'Exponential censoring time')
# method 2: uniform censoring variable
C <- runif(n, 0, 21)
time = pmin(T,C)
status <- 1*(T <= C)
status2 <- 1-status
plot(survfit(Surv(time, status2) ~ 1),
ylab="Survival probability",
main = "Uniform censoring time")
Breslow estimate
- http://support.sas.com/documentation/cdl/en/statug/68162/HTML/default/viewer.htm#statug_lifetest_details03.htm
- Breslow estimate is the exponentiation of the negative Nelson-Aalen estimate of the cumulative hazard function
Survival curves with number at risk at bottom: survminer package
R function survminer::ggsurvplot()
- http://www.sthda.com/english/articles/24-ggpubr-publication-ready-plots/81-ggplot2-easy-way-to-mix-multiple-graphs-on-the-same-page/#mix-table-text-and-ggplot
- http://r-addict.com/2016/05/23/Informative-Survival-Plots.html
Paper examples
Hazard ratio forest plot: ggforest() from survminer
Survival curve with confidence interval
http://www.sthda.com/english/wiki/survminer-r-package-survival-data-analysis-and-visualization
Parametric models and survival function for censored data
Assume the CDF of survival time T is [math]\displaystyle{ F(\cdot) }[/math] and the CDF of the censoring time C is [math]\displaystyle{ G(\cdot) }[/math],
- [math]\displaystyle{ \begin{align} P(T\gt t, \delta=1) &= \int_t^\infty (1-G(s))dF(s), \\ P(T\gt t, \delta=0) &= \int_t^\infty (1-F(s))dG(s) \end{align} }[/math]
- http://www.stat.columbia.edu/~madigan/W2025/notes/survival.pdf#page=23
- http://www.ms.uky.edu/~mai/sta635/LikelihoodCensor635.pdf#page=2 survival function of [math]\displaystyle{ f(T, \delta) }[/math]
- https://web.stanford.edu/~lutian/coursepdf/unit2.pdf#page=3 joint density of [math]\displaystyle{ f(T, \delta) }[/math]
- http://data.princeton.edu/wws509/notes/c7.pdf#page=6
- Special case: T follows Log normal distribution and C follows [math]\displaystyle{ U(0, \xi) }[/math].
R
Parametric models and likelihood function for uncensored data
- Exponential. [math]\displaystyle{ T \sim Exp(\lambda) }[/math]. [math]\displaystyle{ H(t) = \lambda t. }[/math] and [math]\displaystyle{ ln(S(t)) = -H(t) = -\lambda t. }[/math]
- Weibull. [math]\displaystyle{ T \sim W(\lambda,p). }[/math] [math]\displaystyle{ H(t) = \lambda^p t^p. }[/math] and [math]\displaystyle{ ln(-ln(S(t))) = ln(\lambda^p t^p)=const + p ln(t) }[/math].
http://www.math.ucsd.edu/~rxu/math284/slect4.pdf
See also accelerated life models where a set of covariates were used to model survival time.
Survival modeling
Accelerated life models - a direct extension of the classical linear model
http://data.princeton.edu/wws509/notes/c7.pdf and also Kalbfleish and Prentice (1980).
[math]\displaystyle{ log T_i = x_i' \beta + \epsilon_i }[/math] Therefore
- [math]\displaystyle{ T_i = exp(x_i' \beta) T_{0i} }[/math]. So if there are two groups (x=1 and x=0), and [math]\displaystyle{ exp(\beta) = 2 }[/math], it means one group live twice as long as people in another group.
- [math]\displaystyle{ S_1(t) = S_0(t/ exp(x' \beta)) }[/math]. This explains the meaning of accelerated failure-time. Depending on the sign of [math]\displaystyle{ \beta' x }[/math], the time is either accelerated by a constant factor or degraded by a constant factor. If [math]\displaystyle{ exp(\beta)=2 }[/math], the probability that a member in group one (eg treatment) will be alive at age t is exactly the same as the probability that a member in group zero (eg control group) will be alive at age t/2.
- The hazard function [math]\displaystyle{ \lambda_1(t) = \lambda_0(t/exp(x'\beta))/ exp(x'\beta) }[/math]. So if [math]\displaystyle{ exp(\beta)=2 }[/math], at any given age people in group one would be exposed to half the risk of people in group zero half their age.
In applications,
- If the errors are normally distributed, then we obtain a log-normal model for the T. Estimation of this model for censored data by maximum likelihood is known in the econometric literature as a Tobit model.
- If the errors have an extreme value distribution, then T has an exponential distribution. The hazard [math]\displaystyle{ \lambda }[/math] satisfies the log linear model [math]\displaystyle{ \log \lambda_i = x_i' \beta }[/math].
Proportional hazard models
Note PH models is a type of multiplicative hazard rate models [math]\displaystyle{ h(x|Z) = h_0(x)c(\beta' Z) }[/math] where [math]\displaystyle{ c(\beta' Z) = \exp(\beta ' Z) }[/math].
Assumption: Survival curves for two strata (determined by the particular choices of values for covariates) must have hazard functions that are proportional over time (i.e. constant relative hazard over time). Proportional hazards assumption meaning. The ratio of the hazard rates from two individuals with covariate value [math]\displaystyle{ Z }[/math] and [math]\displaystyle{ Z^* }[/math] is a constant function time.
- [math]\displaystyle{ \begin{align} \frac{h(t|Z)}{h(t|Z^*)} = \frac{h_0(t)\exp(\beta 'Z)}{h_0(t)\exp(\beta ' Z^*)} = \exp(\beta' (Z-Z^*)) \mbox{ independent of time} \end{align} }[/math]
Test the assumption
- cox.zph() can be used to test the proportional hazards assumption for a Cox regression model fit.
- Log-log Kaplan-Meier curves and other methods.
- https://stats.idre.ucla.edu/other/examples/asa2/testing-the-proportional-hazard-assumption-in-cox-models/. If the predictor satisfy the proportional hazard assumption then the graph of the survival function versus the survival time should results in a graph with parallel curves, similarly the graph of the log(-log(survival)) versus log of survival time graph should result in parallel lines if the predictor is proportional. This method does not work well for continuous predictor or categorical predictors that have many levels because the graph becomes to “cluttered”.
Weibull and Exponential model to Cox model
- https://socserv.socsci.mcmaster.ca/jfox/Books/Companion/appendix/Appendix-Cox-Regression.pdf. It also includes model diagnostic and all stuff is illustrated in R.
- http://stat.ethz.ch/education/semesters/ss2011/seminar/contents/handout_9.pdf
In summary:
- Weibull distribution (Klein) [math]\displaystyle{ h(t) = p \lambda (\lambda t)^{p-1} }[/math] and [math]\displaystyle{ S(t) = exp(-\lambda t^p) }[/math]. If p >1, then the risk increases over time. If p<1, then the risk decreases over time.
- Note that Weibull distribution has a different parametrization. See http://data.princeton.edu/pop509/ParametricSurvival.pdf#page=2. [math]\displaystyle{ h(t) = \lambda^p p t^{p-1} }[/math] and [math]\displaystyle{ S(t) = exp(-(\lambda t)^p) }[/math]. R and wikipedia also follows this parametrization except that [math]\displaystyle{ h(t) = p t^{p-1}/\lambda^p }[/math] and [math]\displaystyle{ S(t) = exp(-(t/\lambda)^p) }[/math].
- Exponential distribution [math]\displaystyle{ h(t) }[/math] = constant (independent of t). This is a special case of Weibull distribution (p=1).
- Weibull (and also exponential)
distributionregression model is the only case which belongs to both the proportional hazards and the accelerated life families.
- [math]\displaystyle{ \begin{align} \frac{h(x|Z_1)}{h(x|Z_2)} = \frac{h_0(x\exp(-\gamma' Z_1)) \exp(-\gamma ' Z_1)}{h_0(x\exp(-\gamma' Z_2)) \exp(-\gamma ' Z_2)} = \frac{(a/b)\left(\frac{x \exp(-\gamma ' Z_1)}{b}\right)^{a-1}\exp(-\gamma ' Z_1)}{(a/b)\left(\frac{x \exp(-\gamma ' Z_2)}{b}\right)^{a-1}\exp(-\gamma ' Z_2)} \quad \mbox{which is independent of time x} \end{align} }[/math]
- Using the Weibull baseline hazard is the only circumstance under which the model satisfies both the proportional hazards, and accelerated failure time models
- If X is exponential distribution with mean [math]\displaystyle{ b }[/math], then X^(1/a) follows Weibull(a, b). See Exponential distribution and Weibull distribution.
- Derivation of mean and variance of Weibull distribution.
| f(t)=h(t)*S(t) | h(t) | S(t) | Mean | |
|---|---|---|---|---|
| Exponential (Klein p37) | [math]\displaystyle{ \lambda \exp(-\lambda t) }[/math] | [math]\displaystyle{ \lambda }[/math] | [math]\displaystyle{ \exp(-\lambda t) }[/math] | [math]\displaystyle{ 1/\lambda }[/math] |
| Weibull (Klein, wikipedia) | [math]\displaystyle{ p\lambda t^{p-1}\exp(-\lambda t^p) }[/math] | [math]\displaystyle{ p\lambda t^{p-1} }[/math] | [math]\displaystyle{ exp(-\lambda t^p) }[/math] | [math]\displaystyle{ \frac{\Gamma(1+1/p)}{\lambda^{1/p}} }[/math] |
| Exponential (R) | [math]\displaystyle{ \lambda \exp(-\lambda t) }[/math], [math]\displaystyle{ \lambda }[/math] is rate | [math]\displaystyle{ \lambda }[/math] | [math]\displaystyle{ \exp(-\lambda t) }[/math] | [math]\displaystyle{ 1/\lambda }[/math] |
| Weibull (R, wikipedia) | [math]\displaystyle{ \frac{a}{b}\left(\frac{x}{b}\right)^{a-1} \exp(-(\frac{x}{b})^a) }[/math], [math]\displaystyle{ a }[/math] is shape, and [math]\displaystyle{ b }[/math] is scale |
[math]\displaystyle{ \frac{a}{b}\left(\frac{x}{b}\right)^{a-1} }[/math] | [math]\displaystyle{ \exp(-(\frac{x}{b})^a) }[/math] | [math]\displaystyle{ b\Gamma(1+1/a) }[/math] |
- Accelerated failure-time model. Let [math]\displaystyle{ Y=\log(T)=\mu + \gamma'Z + \sigma W }[/math]. Then the survival function of [math]\displaystyle{ T }[/math] at the covariate Z,
- [math]\displaystyle{ \begin{align} S_T(t|Z) &= P(T \gt t |Z) \\ &= P(Y \gt \ln t|Z) \\ &= P(\mu + \sigma W \gt \ln t-\gamma' Z | Z) \\ &= P(e^{\mu + \sigma W} \gt t\exp(-\gamma'Z) | Z) \\ &= S_0(t \exp(-\gamma'Z)). \end{align} }[/math]
where [math]\displaystyle{ S_0(t) }[/math] denote the survival function T when Z=0. Since [math]\displaystyle{ h(t) = -\partial \ln (S(t)) }[/math], the hazard function of T with a covariate value Z is related to a baseline hazard rate [math]\displaystyle{ h_0 }[/math] by (p56 Klein)
- [math]\displaystyle{ \begin{align} h(t|Z) = h_0(t\exp(-\gamma' Z)) \exp(-\gamma ' Z) \end{align} }[/math]
> mean(rexp(1000)^(1/2)) [1] 0.8902948 > mean(rweibull(1000, 2, 1)) [1] 0.8856265 > mean((rweibull(1000, 2, scale=4)/4)^2) [1] 1.008923
Graphical way to check Weibull, AFT, PH
http://stat.ethz.ch/education/semesters/ss2011/seminar/contents/handout_9.pdf#page=40
CDF follows Unif(0,1)
https://stats.stackexchange.com/questions/161635/why-is-the-cdf-of-a-sample-uniformly-distributed
Take the Exponential distribution for example
stem(pexp(rexp(1000))) stem(pexp(rexp(10000)))
Another example is from simulating survival time. Note that this is exactly Bender et al 2005 approach. See also the simsurv (newer) and survsim (older) packages.
set.seed(100) #Define the following parameters outlined in the step: n = 1000 beta_0 = 0.5 beta_1 = -1 beta_2 = 1 b = 1.6 #This will be changed later as mentioned in Step 5 of documentation #Step 1 x_1<-rbinom(n, 1, 0.25) x_2<-rbinom(n, 1, 0.7) #Step 2 U<-runif(n, 0,1) T<-(-log(U)*exp(-(beta_0+beta_1*x_1+beta_2*x_2))) #Eqn (5) Fn <- ecdf(T) # https://stat.ethz.ch/R-manual/R-devel/library/stats/html/ecdf.html # verify F(T) or 1-F(T) ~ U(0, 1) hist(Fn(T)) # look at the plot of survival probability vs time plot(T, 1 - Fn(T))
Simulate survival data
Note that status = 1 means an event (e.g. death) happened; Ti <= Ci. That is, the status variable used in R/Splus means the death indicator.
y <- rexp(10) cen <- runif(10) status <- ifelse(cen < .7, 1, 0)
- How much power/accuracy is lost by using the Cox model instead of Weibull model when both model are correct? [math]\displaystyle{ h(t|x)=\lambda=e^{3x+1} = h_0(t)e^{\beta x} }[/math] where [math]\displaystyle{ h_0(t)=e^1, \beta=3 }[/math].
n <- 30
x <- scale(1:n, TRUE, TRUE) # create covariates (standardized)
# the original example does not work on large 'n'
myrates <- exp(3*x+1)
set.seed(1234)
y <- rexp(n, rate = myrates) # generates the r.v.
cen <- rexp(n, rate = 0.5 ) # E(cen)=1/rate
ycen <- pmin(y, cen)
di <- as.numeric(y <= cen)
survreg(Surv(ycen, di)~x, dist="weibull")$coef[2] # -3.080125
coxph(Surv(ycen, di)~x)$coef # 2.457466
# no censor
survreg(Surv(y,rep(1, n))~x,dist="weibull")$coef[2] # -3.137603
survreg(Surv(y,rep(1, n))~x,dist="exponential")$coef[2] # -3.143095
coxph(Surv(y,rep(1, n))~x)$coef # 2.717794
# See the pdf note for the rest of code
- Intercept in survreg for the exponential distribution. http://www.stat.columbia.edu/~madigan/W2025/notes/survival.pdf#page=25.
- [math]\displaystyle{ \begin{align} \lambda = exp(-intercept) \end{align} }[/math]
> futime <- rexp(1000, 5) > survreg(Surv(futime,rep(1,1000))~1,dist="exponential")$coef (Intercept) -1.618263 > exp(1.618263) [1] 5.044321
- Intercept and scale in survreg for a Weibull distribution. http://www.stat.columbia.edu/~madigan/W2025/notes/survival.pdf#page=28.
- [math]\displaystyle{ \begin{align} \gamma &= 1/scale \\ \alpha &= exp(-(Intercept)*\gamma) \end{align} }[/math]
> survreg(Surv(futime,rep(1,1000))~1,dist="weibull") Call: survreg(formula = Surv(futime, rep(1, 1000)) ~ 1, dist = "weibull") Coefficients: (Intercept) -1.639469 Scale= 1.048049 Loglik(model)= 620.1 Loglik(intercept only)= 620.1 n= 1000
- rsurv() function from the ipred package
- Use Weibull distribution to model survival data. We assume the shape is constant across subjects. We then allow the scale to vary across subjects. For subject [math]\displaystyle{ i }[/math] with covariate [math]\displaystyle{ X_i }[/math], [math]\displaystyle{ \log(scale_i) }[/math] = [math]\displaystyle{ \beta ' X_i }[/math]. Note that if we want to make the [math]\displaystyle{ \beta }[/math] sign to be consistent with the Cox model, we want to use [math]\displaystyle{ \log(scale_i) }[/math] = [math]\displaystyle{ -\beta ' X_i }[/math] instead.
- http://sas-and-r.blogspot.com/2010/03/example-730-simulate-censored-survival.html. Assuming shape=1 in the Weibull distribution, then the hazard function can be expressed as a proportional hazard model
[math]\displaystyle{ h(t|x) = 1/scale = \frac{1}{\lambda/e^{\beta 'x}} = \frac{e^{\beta ' x}}{\lambda} = h_0(t) \exp(\beta' x) }[/math]
n = 10000 beta1 = 2; beta2 = -1 lambdaT = .002 # baseline hazard lambdaC = .004 # hazard of censoring set.seed(1234) x1 = rnorm(n,0) x2 = rnorm(n,0) # true event time T = rweibull(n, shape=1, scale=lambdaT*exp(-beta1*x1-beta2*x2)) # No censoring event2 <- rep(1, length(T)) coxph(Surv(T, event2)~ x1 + x2) # coef exp(coef) se(coef) z p # x1 1.9982 7.3761 0.0188 106.1 <2e-16 # x2 -1.0020 0.3671 0.0127 -79.1 <2e-16 # # Likelihood ratio test=15556 on 2 df, p=0 # n= 10000, number of events= 10000 # Censoring C = rweibull(n, shape=1, scale=lambdaC) #censoring time time = pmin(T,C) #observed time is min of censored and true event = time==T # set to 1 if event is observed coxph(Surv(time, event)~ x1 + x2) # coef exp(coef) se(coef) z p # x1 2.0104 7.4662 0.0225 89.3 <2e-16 # x2 -0.9921 0.3708 0.0155 -63.9 <2e-16 # # Likelihood ratio test=11321 on 2 df, p=0 # n= 10000, number of events= 6002
- https://stats.stackexchange.com/questions/135124/how-to-create-a-toy-survival-time-to-event-data-with-right-censoring (Bender's inverse probability method)
# N = sample size
# lambda = scale parameter in h0()
# rho = shape parameter in h0()
# beta = fixed effect parameter
# rateC = rate parameter of the exponential distribution of C
simulWeib <- function(N, lambda, rho, beta, rateC)
{
# covariate --> N Bernoulli trials
x <- sample(x=c(0, 1), size=N, replace=TRUE, prob=c(0.5, 0.5))
# Weibull latent event times
v <- runif(n=N)
Tlat <- (- log(v) / (lambda * exp(x * beta)))^(1 / rho)
# censoring times
C <- rexp(n=N, rate=rateC)
# follow-up times and event indicators
time <- pmin(Tlat, C)
status <- as.numeric(Tlat <= C)
# data set
data.frame(id=1:N,
time=time,
status=status,
x=x)
}
# Test
set.seed(1234)
betaHat <- rep(NA, 1e3)
for(k in 1:1e3)
{
dat <- simulWeib(N=100, lambda=0.01, rho=1, beta=-0.6, rateC=0.001)
fit <- coxph(Surv(time, status) ~ x, data=dat)
betaHat[k] <- fit$coef
}
> mean(betaHat)
[1] -0.6085473
- Generating survival times to simulate Cox proportional hazards models Bender et al 2005
- survsim package and the paper on JSS. See this post.
- simsurv package (new, 2 vignettes).
- Get a desired percentage of censored observations in a simulation of Cox PH Model. The answer is based on Bender et al 2005. Generating survival times to simulate Cox proportional hazards models. Statistics in Medicine 24: 1713–1723. The censoring time is fixed and the distribution of the censoring indicator is following the binomial. In fact, when we simulate survival data with a predefined censoring rate, we can pretend the survival time is already censored and only care about the censoring/status variable to make sure the censoring rate is controlled.
- (Search github) Using inverse CDF [math]\displaystyle{ \lambda = exp(\beta' x), \; S(t)= \exp(-\lambda t) = \exp(-t e^{\beta' x}) \sim Unif(0,1) }[/math]
Predefined censoring rates
Simulating survival data with predefined censoring rates for proportional hazards models
Cross validation
- Cross validation in survival analysis by Verweij & van Houwelingen, Stat in medicine 1993.
- Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Simon et al, Brief Bioinform. 2011
Survival rate
- Disease-free survival (DFS): the period after curative treatment [disease eliminated] when no disease can be detected
- Progression-free survival (PFS), overall survival (OS). PFS is the length of time during and after the treatment of a disease, such as cancer, that a patient lives with the disease but it does not get worse. See an use at NCI-MATCH trial.
- Time to progression: The length of time from the date of diagnosis or the start of treatment for a disease until the disease starts to get worse or spread to other parts of the body. In a clinical trial, measuring the time to progression is one way to see how well a new treatment works. Also called TTP.
- Metastasis-free survival (MFS) time: the period until metastasis is detected
- Understanding Statistics Used to Guide Prognosis and Evaluate Treatment (DFS & PFS rate)
Books
- Survival Analysis, A Self-Learning Text by Kleinbaum, David G., Klein, Mitchel
- Applied Survival Analysis Using R by Moore, Dirk F.
- Regression Modeling Strategies by Harrell, Frank
- Regression Methods in Biostatistics by Vittinghoff, E., Glidden, D.V., Shiboski, S.C., McCulloch, C.E.
- https://tbrieder.org/epidata/course_reading/e_tableman.pdf
- Survival Analysis: Models and Applications by Xian Liu
HER2-positive breast cancer
- https://www.mayoclinic.org/breast-cancer/expert-answers/FAQ-20058066
- https://en.wikipedia.org/wiki/Trastuzumab (antibody, injection into a vein or under the skin)
Cox proportional hazards model
Let Yi denote the observed time (either censoring time or event time) for subject i, and let Ci be the indicator that the time corresponds to an event (i.e. if Ci = 1 the event occurred and if Ci = 0 the time is a censoring time). The hazard function for the Cox proportional hazard model has the form
[math]\displaystyle{ \lambda(t|X) = \lambda_0(t)\exp(\beta_1X_1 + \cdots + \beta_pX_p) = \lambda_0(t)\exp(X \beta^\prime). }[/math]
This expression gives the hazard at time t for an individual with covariate vector (explanatory variables) X. Based on this hazard function, a partial likelihood (defined on hazard function) can be constructed from the datasets as
[math]\displaystyle{ L(\beta) = \prod_{i:C_i=1}\frac{\theta_i}{\sum_{j:Y_j\ge Y_i}\theta_j}, }[/math]
where θj = exp(Xj β′) and X1, ..., Xn are the covariate vectors for the n independently sampled individuals in the dataset (treated here as column vectors). This pdf or this note give a toy example
The corresponding log partial likelihood is
[math]\displaystyle{ \ell(\beta) = \sum_{i:C_i=1} \left(X_i \beta^\prime - \log \sum_{j:Y_j\ge Y_i}\theta_j\right). }[/math]
This function can be maximized over β to produce maximum partial likelihood estimates of the model parameters.
The partial score function is [math]\displaystyle{ \ell^\prime(\beta) = \sum_{i:C_i=1} \left(X_i - \frac{\sum_{j:Y_j\ge Y_i}\theta_jX_j}{\sum_{j:Y_j\ge Y_i}\theta_j}\right), }[/math]
and the Hessian matrix of the partial log likelihood is
[math]\displaystyle{ \ell^{\prime\prime}(\beta) = -\sum_{i:C_i=1} \left(\frac{\sum_{j:Y_j\ge Y_i}\theta_jX_jX_j^\prime}{\sum_{j:Y_j\ge Y_i}\theta_j} - \frac{\sum_{j:Y_j\ge Y_i}\theta_jX_j\times \sum_{j:Y_j\ge Y_i}\theta_jX_j^\prime}{[\sum_{j:Y_j\ge Y_i}\theta_j]^2}\right). }[/math]
Using this score function and Hessian matrix, the partial likelihood can be maximized using the Newton-Raphson algorithm. The inverse of the Hessian matrix, evaluated at the estimate of β, can be used as an approximate variance-covariance matrix for the estimate, and used to produce approximate standard errors for the regression coefficients.
If X is age, then the coefficient is likely >0. If X is some treatment, then the coefficient is likely <0.
Compare the partial likelihood to the full likelihood
http://math.ucsd.edu/~rxu/math284/slect5.pdf#page=10
z-column (Wald statistic) from R's coxph()
- https://socialsciences.mcmaster.ca/jfox/Books/Companion/appendix/Appendix-Cox-Regression.pdf#page=6 The ratio of each regression coefficient to its standard error, a Wald statistic which is asymptotically standard normal under the hypothesis that the corresponding β is 0.
- http://dni-institute.in/blogs/cox-regression-interpret-result-and-predict/
How exactly can the Cox-model ignore exact times?
The Cox model does not depend on the times itself, instead it only needs an ordering of the events.
library(survival) survfit(Surv(time, status) ~ x, data = aml) fit <- coxph(Surv(time, status) ~ x, data = aml) coef(fit) # 0.9155326 min(diff(sort(unique(aml$time)))) # 1 # Shift survival time for some obs but keeps the same order # make sure we choose obs (n=20 not works but n=21 works) with twins rbind(order(aml$time), sort(aml$time), aml$time[order(aml$time)]) # [,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8] [,9] [,10] [,11] [,12] [,13] [,14] [,15] [,16] # [1,] 12 13 14 15 1 16 2 3 17 4 5 18 19 6 20 7 # [2,] 5 5 8 8 9 12 13 13 16 18 23 23 27 28 30 31 # [3,] 5 5 8 8 9 12 13 13 16 18 23 23 27 28 30 31 # [,17] [,18] [,19] [,20] [,21] [,22] [,23] # [1,] 21 8 22 9 23 10 11 # [2,] 33 34 43 45 45 48 161 # [3,] 33 34 43 45 45 48 161 aml$time2 <- aml$time aml$time2[order(aml$time)[1:21]] <- aml$time[order(aml$time)[1:21]] - .9 fit2 <- coxph(Surv(time2, status) ~ x, data = aml); fit2 coef(fit2) # 0.9155326 coef(fit) == coef(fit2) # TRUE aml$time3 <- aml$time aml$time3[order(aml$time)[1:20]] <- aml$time[order(aml$time)[1:20]] - .9 fit3 <- coxph(Surv(time3, status) ~ x, data = aml); fit3 coef(fit3) # 0.8891567 coef(fit) == coef(fit3) # FALSE
Partial likelihood when there are ties; hypothesis testing: Likelihood Ratio Test, Wald Test & Score Test
http://math.ucsd.edu/~rxu/math284/slect5.pdf#page=29
In R's coxph(): Nearly all Cox regression programs use the Breslow method by default, but not this one. The Efron approximation is used as the default here, it is more accurate when dealing with tied death times, and is as efficient computationally.
http://sfb649.wiwi.hu-berlin.de/fedc_homepage/xplore/tutorials/xaghtmlnode28.html (include the case when there is a partition of parameters). The formulas for 3 tests are also available on Appendix B of Klein book.
The following code does not test for models. But since there is only one coefficient, the results are the same. If there is more than one variable, we can use anova(model1, model2) to run LRT.
library(KMsurv) # No ties. Section 8.2 data(btrial) str(btrial) # 'data.frame': 45 obs. of 3 variables: # $ time : int 19 25 30 34 37 46 47 51 56 57 ... # $ death: int 1 1 1 1 1 1 1 1 1 1 ... # $ im : int 1 1 1 1 1 1 1 1 1 1 ... table(subset(btrial, death == 1)$time) # death time is unique coxph(Surv(time, death) ~ im, data = btrial) # coef exp(coef) se(coef) z p # im 0.980 2.665 0.435 2.25 0.024 # Likelihood ratio test=4.45 on 1 df, p=0.03 # n= 45, number of events= 24 # Ties, Section 8.3 data(kidney) str(kidney) # 'data.frame': 119 obs. of 3 variables: # $ time : num 1.5 3.5 4.5 4.5 5.5 8.5 8.5 9.5 10.5 11.5 ... # $ delta: int 1 1 1 1 1 1 1 1 1 1 ... # $ type : int 1 1 1 1 1 1 1 1 1 1 ... table(subset(kidney, delta == 1)$time) # 0.5 1.5 2.5 3.5 4.5 5.5 6.5 8.5 9.5 10.5 11.5 15.5 16.5 18.5 23.5 26.5 # 6 1 2 2 2 1 1 2 1 1 1 2 1 1 1 1 # Default: Efron method coxph(Surv(time, delta) ~ type, data = kidney) # coef exp(coef) se(coef) z p # type -0.613 0.542 0.398 -1.54 0.12 # Likelihood ratio test=2.41 on 1 df, p=0.1 # n= 119, number of events= 26 summary(coxph(Surv(time, delta) ~ type, data = kidney)) # n= 119, number of events= 26 # coef exp(coef) se(coef) z Pr(>|z|) # type -0.6126 0.5420 0.3979 -1.539 0.124 # # exp(coef) exp(-coef) lower .95 upper .95 # type 0.542 1.845 0.2485 1.182 # # Concordance= 0.497 (se = 0.056 ) # Rsquare= 0.02 (max possible= 0.827 ) # Likelihood ratio test= 2.41 on 1 df, p=0.1 # Wald test = 2.37 on 1 df, p=0.1 # Score (logrank) test = 2.44 on 1 df, p=0.1 # Breslow method summary(coxph(Surv(time, delta) ~ type, data = kidney, ties = "breslow")) # n= 119, number of events= 26 # coef exp(coef) se(coef) z Pr(>|z|) # type -0.6182 0.5389 0.3981 -1.553 0.12 # # exp(coef) exp(-coef) lower .95 upper .95 # type 0.5389 1.856 0.247 1.176 # # Concordance= 0.497 (se = 0.056 ) # Rsquare= 0.02 (max possible= 0.827 ) # Likelihood ratio test= 2.45 on 1 df, p=0.1 # Wald test = 2.41 on 1 df, p=0.1 # Score (logrank) test = 2.49 on 1 df, p=0.1 # Discrete/exact method summary(coxph(Surv(time, delta) ~ type, data = kidney, ties = "exact")) # coef exp(coef) se(coef) z Pr(>|z|) # type -0.6294 0.5329 0.4019 -1.566 0.117 # # exp(coef) exp(-coef) lower .95 upper .95 # type 0.5329 1.877 0.2424 1.171 # # Rsquare= 0.021 (max possible= 0.795 ) # Likelihood ratio test= 2.49 on 1 df, p=0.1 # Wald test = 2.45 on 1 df, p=0.1 # Score (logrank) test = 2.53 on 1 df, p=0.1
Hazard (function) and survival function
A hazard is the rate at which events happen, so that the probability of an event happening in a short time interval is the length of time multiplied by the hazard.
[math]\displaystyle{ h(t) = \lim_{\Delta t \to 0} \frac{P(t \leq T \lt t+\Delta t|T \geq t)}{\Delta t} = \frac{f(t)}{S(t)} = -\partial{ln[S(t)]} }[/math]
Therefore
[math]\displaystyle{ H(x) = \int_0^x h(u) d(u) = -ln[S(x)]. }[/math]
or
[math]\displaystyle{ S(x) = e^{-H(x)} }[/math]
Hazards (or probability of hazards) may vary with time, while the assumption in proportional hazard models for survival is that the hazard is a constant proportion.
Examples:
- If h(t)=c, S(t) is exponential. f(t) = c exp(-ct). The mean is 1/c.
- If [math]\displaystyle{ \log h(t) = c + \rho t }[/math], S(t) is Gompertz distribution.
- If [math]\displaystyle{ \log h(t)=c + \rho \log (t) }[/math], S(t) is Weibull distribution.
- For Cox regression, the survival function can be shown to be [math]\displaystyle{ S(t|X) = S_0(t) ^ {\exp(X\beta)} }[/math].
- [math]\displaystyle{ \begin{align} S(t|X) &= e^{-H(t)} = e^{-\int_0^t h(u|X)du} \\ &= e^{-\int_0^t h_0(u) exp(X\beta) du} \\ &= e^{-\int_0^t h_0(u) du \cdot exp(X \beta)} \\ &= S_0(t)^{exp(X \beta)} \end{align} }[/math]
Alternatively,
- [math]\displaystyle{ \begin{align} S(t|X) &= e^{-H(t)} = e^{-\int_0^t h(u|X)du} \\ &= e^{-\int_0^t h_0(u) exp(X\beta) du} \\ &= e^{-H_0(t) \cdot exp(X \beta)} \end{align} }[/math]
where the cumulative baseline hazard at time t, [math]\displaystyle{ H_0(t) }[/math], is commonly estimated through the non-parametric Breslow estimator.
Check the proportional hazard (constant HR over time) assumption by cox.zph()
- https://www.rdocumentation.org/packages/survival/versions/2.41-2/topics/cox.zph
- http://rstudio-pubs-static.s3.amazonaws.com/5896_8f0fed2ccbbd42489276e554a05af87e.html
Extract p-values
fit <- coxph(Surv(futime, fustat) ~ age, data = ovarian) # method 1: beta <- coef(fit) se <- sqrt(diag(vcov(fit))) 1 - pchisq((beta/se)^2, 1) # method 2: https://www.biostars.org/p/65315/ coef(summary(fit))[, "Pr(>|z|)"]
Expectation of life & expected future lifetime
- The average lifetime is the same as the area under the survival curve.
- [math]\displaystyle{ \begin{align} \mu &= \int_0^\infty t f(t) dt \\ &= \int_0^\infty S(t) dt \end{align} }[/math]
by integrating by parts making use of the fact that -f(t) is the derivative of S(t), which has limits S(0)=1 and [math]\displaystyle{ S(\infty)=0 }[/math]. The average lifetime may not be bounded if you have censored data, there's censored observations that last beyond your last recorded death.
- [math]\displaystyle{ \frac{1}{S(t_0)} \int_0^{\infty} t\,f(t_0+t)\,dt = \frac{1}{S(t_0)} \int_{t_0}^{\infty} S(t)\,dt, }[/math]
Hazard Ratio/Relative Risk
A hazard ratio is often reported as a “reduction in risk of death or progression” – This risk reduction is calculated as 1 minus the Hazard Ratio (exp^beta), e.g., HR of 0.84 is equal to a 16% reduction in risk. See www.time4epi.com and stackexchange.com.
Another example (John Fox) is assuming Y ~ age + prio + others.
- If exp(beta_age) = 0.944. It means an additional year of age reduces the hazard by a factor of .944 on average, or (1-.944)*100 = 5.6 percent.
- If exp(beta_prio) = 1.096, it means each prior conviction increases the hazard by a factor of 1.096, or 9.6 percent.
Hazard ratio is not the same as the relative risk ratio. See medicine.ox.ac.uk.
Interpreting risks and ratios in therapy trials from australianprescriber.com is useful too.
For two groups that differ only in treatment condition, the ratio of the hazard functions is given by [math]\displaystyle{ e^\beta }[/math], where [math]\displaystyle{ \beta }[/math] is the estimate of treatment effect derived from the regression model. See here.
Compute ratio ratios from coxph() in R (Hint: exp(beta)).
Prognostic index is defined on http://www.math.ucsd.edu/~rxu/math284/slect6.pdf#page=2.
Basics of the Cox proportional hazards model. Good prognostic factor (b<0 or HR<1) and bad prognostic factor (b>0 or HR>1).
Variable selection: variables were retained in the prediction models if they had a hazard ratio of <0.85 or >1.15 (for binary variables) and were statistically significant at the 0.01 level. see Development and validation of risk prediction equations to estimate survival in patients with colorectal cancer: cohort study.
Hazard Ratio and death probability
https://en.wikipedia.org/wiki/Hazard_ratio#The_hazard_ratio_and_survival
Suppose S0(t)=.2 (20% survived at time t) and the hazard ratio (hr) is 2 (a group has twice the chance of dying than a comparison group), then (Cox model is assumed)
- S1(t)=S0(t)hr = .22 = .04 (4% survived at t)
- The corresponding death probabilities are 0.8 and 0.96.
- If a subject is exposed to twice the risk of a reference subject at every age, then the probability that the subject will be alive at any given age is the square of the probability that the reference subject (covariates = 0) would be alive at the same age. See p10 of this lecture notes.
- exp(x*beta) is the relative risk associated with covariate value x.
Hazard Ratio Forest Plot
The forest plot quickly summarizes the hazard ratio data across multiple variables –If the line crosses the 1.0 value, the hazard ratio is not significant and there is no clear advantage for either arm.
Piece-wise constant baseline hazard model, Poisson model and Breslow estimate
- https://en.wikipedia.org/wiki/Proportional_hazards_model#Relationship_to_Poisson_models
- http://data.princeton.edu/wws509/notes/c7s4.html
- It has been implemented in the biospear package (poissonize.R) with the 'grplasso' package for group-lasso method. We implemented a Poisson model over two-month intervals, corresponding to a piecewise constant hazard model which approximates rather well the Breslow estimator in the Cox model.
- http://r.789695.n4.nabble.com/exponential-proportional-hazard-model-td805536.html
- https://www.demogr.mpg.de/papers/technicalreports/tr-2010-003.pdf
- Does Cox Regression have an underlying Poisson distribution?
- https://rdrr.io/cran/JM/man/piecewiseExp.ph.html
- https://rdrr.io/cran/pch/man/pchreg.html
- Survival Analysis via Hazard Based Modeling and Generalized Linear Models
- https://www.rdocumentation.org/packages/mgcv/versions/1.8-23/topics/cox.pht
Estimate baseline hazard [math]\displaystyle{ h_0(t) }[/math], Breslow cumulative baseline hazard [math]\displaystyle{ H_0(t) }[/math], baseline survival function [math]\displaystyle{ S_0(t) }[/math] and the survival function [math]\displaystyle{ S(t) }[/math]
Google: how to estimate baseline hazard rate
- survfit.object has print(), plot(), lines(), and points() methods. It returns a list with components
- n
- time
- n.risk
- n.event
- n.censor
- surv [S_0(t)]
- cumhaz [ same as -log(surv)]
- upper
- lower
- n.all
- Terry Therneau: The baseline survival, which is the survival for a hypothetical subject with all covariates=0, may be useful mathematical shorthand when writing a book but I cannot think of a single case where the resulting curve would be of any practical interest in medical data.
- http://www.math.ucsd.edu/~rxu/math284/slect6.pdf#page=4 Breslow Estimator for cumulative baseline hazard at a time t and Kalbfleisch/Prentice Estimator
- When there are no covariates, the Breslow’s estimate reduces to the Fleming-Harrington (Nelson-Aalen) estimate, and K/P reduces to KM.
- stackexchange.com and cumulative and non-cumulative baseline hazard
- (newbie) Cox Baseline Hazard There are two methods of calculating the baseline survival, the default one gives the baseline hazard estimator you want. It is attributed to Aalen, Breslow, or Peto (see the next item). An example: https://stats.idre.ucla.edu/r/examples/asa/r-applied-survival-analysis-ch-2/.
- survfit.coxph(formula, newdata, type, ...)
- newdata: Default is the mean of the covariates used in the coxph fit
- type = "aalen", "efron", or "kalbfleisch-prentice". The default is to match the computation used in the Cox model. The Nelson-Aalen-Breslow estimate for ties='breslow', the Efron estimate for ties='efron' and the Kalbfleisch-Prentice estimate for a discrete time model ties='exact'. Variance estimates are the Aalen-Link-Tsiatis, Efron, and Greenwood. The default will be the Efron estimate for ties='efron' and the Aalen estimate otherwise.
- Nelson-Aalen estimator in R. The easiest way to get the Nelson-Aalen estimator is
basehaz(coxph(Surv(time,status)~1,data=aml))
because the (Breslow) hazard estimator for a Cox model reduces to the Nelson-Aalen estimator when there are no covariates. You can also compute it from information returned by survfit().
fit <- survfit(Surv(time, status) ~ 1, data = aml) cumsum(fit$n.event/fit$n.risk) # the Nelson-Aalen estimator for the times given by fit$times -log(fit$surv) # cumulative hazard
Manually compute
Breslow estimator of the baseline cumulative hazard rate reduces to the Nelson-Aalen estimator when there are no covariates present; see p283 of Klein 2003.
- [math]\displaystyle{ \begin{align} \hat{H}_0(t) &= \sum_{t_i \le t} \frac{d_i}{W(t_i;b)}, \\ W(t_i;b) &= \sum_{j \in R(t_i)} \exp(\sum_{k=1}^p b_k z_{jk}) \end{align} }[/math]
where [math]\displaystyle{ t_1 \lt t_2 \lt \cdots \lt t_D }[/math] denotes the distinct death times and [math]\displaystyle{ d_i }[/math] be the number of deaths at time [math]\displaystyle{ t_i }[/math]. The estimator of the baseline survival function [math]\displaystyle{ S_0(t) = \exp [-H_0(t)] }[/math] is given by [math]\displaystyle{ \hat{S}_0(t) = \exp [-\hat{H}_0(t)] }[/math]. Below we use the formula to compute the cumulative hazard (and survival function) and compare them with the result obtained using R's built-in functions. The following code is a modification of the snippet from the post Breslow cumulative hazard and basehaz().
bhaz <- function(beta, time, status, x) {
# time can be duplicated
# x (covariate) must be continuous
data <- data.frame(time,status,x)
data <- data[order(data$time), ]
dt <- unique(data$time)
k <- length(dt)
risk <- exp(data.matrix(data[,-c(1:2)]) %*% beta)
h <- rep(0,k)
for(i in 1:k) {
h[i] <- sum(data$status[data$time==dt[i]]) / sum(risk[data$time>=dt[i]])
}
return(data.frame(h, dt))
}
# Example 1 'ovarian' which has unique survival time
all(table(ovarian$futime) == 1) # TRUE
fit <- coxph(Surv(futime, fustat) ~ age, data = ovarian)
# 1. compute the cumulative baseline hazard
# 1.1 manually using Breslow estimator at the observed time
h0 <- bhaz(fit$coef, ovarian$futime, ovarian$fustat, ovarian$age)
H0 <- cumsum(h0$h)
# 1.2 use basehaz (always compute at the observed time)
# since we consider the baseline, we need to use centered=FALSE
H0.bh <- basehaz(fit, centered = FALSE)
cbind(H0, h0$dt, H0.bh)
range(abs(H0 - H0.bh$hazard)) # [1] 6.352747e-22 5.421011e-20
# 2. compute the estimation of the survival function
# 2.1 manually using Breslow estimator at t = observed time (one dim, sorted)
# and observed age (another dim):
# S(t) = S0(t) ^ exp(bx) = exp(-H0(t)) ^ exp(bx)
S1 <- outer(exp(-H0), exp(coef(fit) * ovarian$age), "^")
dim(S1) # row = times, col = age
# 2.2 How about considering times not at observed (e.g. h0$dt - 10)?
# Let's look at the hazard rate
newtime <- h0$dt - 10
H0 <- sapply(newtime, function(tt) sum(h0$h[h0$dt <= tt]))
S2 <- outer(exp(-H0), exp(coef(fit) * ovarian$age), "^")
dim(S2) # row = newtime, col = age
# 2.3 use summary() and survfit() to obtain the estimation of the survival
S3 <- summary(survfit(fit, data.frame(age = ovarian$age)), times = h0$dt)$surv
dim(S3) # row = times, col = age
range(abs(S1 - S3)) # [1] 2.117244e-17 6.544321e-12
# 2.4 How about considering times not at observed (e.g. h0$dt - 10)?
# Note that we cannot put times larger than the observed
S4 <- summary(survfit(fit, data.frame(age = ovarian$age)), times = newtime)$surv
range(abs(S2 - S4)) # [1] 0.000000e+00 6.544321e-12
# Example 2 'kidney' which has duplicated time fit <- coxph(Surv(time, status) ~ age, data = kidney) # manually compute the breslow cumulative baseline hazard # at the observed time h0 <- with(kidney, bhaz(fit$coef, time, status, age)) H0 <- cumsum(h0$h) # use basehaz (always compute at the observed time) # since we consider the baseline, we need to use centered=FALSE H0.bh <- basehaz(fit, centered = FALSE) head(cbind(H0, h0$dt, H0.bh)) range(abs(H0 - H0.bh$hazard)) # [1] 0.000000000 0.005775414 # manually compute the estimation of the survival function # at t = observed time (one dim, sorted) and observed age (another dim): # S(t) = S0(t) ^ exp(bx) = exp(-H0(t)) ^ exp(bx) S1 <- outer(exp(-H0), exp(coef(fit) * kidney$age), "^") dim(S1) # row = times, col = age # How about considering times not at observed (h0$dt - 1)? # Let's look at the hazard rate newtime <- h0$dt - 1 H0 <- sapply(newtime, function(tt) sum(h0$h[h0$dt <= tt])) S2 <- outer(exp(-H0), exp(coef(fit) * kidney$age), "^") dim(S2) # row = newtime, col = age # use summary() and survfit() to obtain the estimation of the survival S3 <- summary(survfit(fit, data.frame(age = kidney$age)), times = h0$dt)$surv dim(S3) # row = times, col = age range(abs(S1 - S3)) # [1] 0.000000000 0.002783715 # How about considering times not at observed (h0$dt - 1)? # We cannot put times larger than the observed S4 <- summary(survfit(fit, data.frame(age = kidney$age)), times = newtime)$surv range(abs(S2 - S4)) # [1] 0.000000000 0.002783715
- basehaz() (an alias for survfit) from stackexchange.com and here. basehaz() has a parameter centered which by default is TRUE. Actually basehaz() gives cumulative hazard H(t). See here. That is, exp(-basehaz(fit)$hazard) is the same as summary(survfit(fit))$surv. basehaz() function is not useful.
fit <- coxph(Surv(futime, fustat) ~ age, data = ovarian)
> fit
Call:
coxph(formula = Surv(futime, fustat) ~ age, data = ovarian)
coef exp(coef) se(coef) z p
age 0.1616 1.1754 0.0497 3.25 0.0012
Likelihood ratio test=14.3 on 1 df, p=0.000156
n= 26, number of events= 12
# Note the default 'centered = TRUE' for basehaz()
> exp(-basehaz(fit)$hazard) # exp(-cumulative hazard)
[1] 0.9880206 0.9738738 0.9545899 0.9334790 0.8973620 0.8624781 0.8243117
[8] 0.8243117 0.8243117 0.7750981 0.7750981 0.7244924 0.6734146 0.6734146
[15] 0.5962187 0.5204807 0.5204807 0.5204807 0.5204807 0.5204807 0.5204807
[22] 0.5204807 0.5204807 0.5204807 0.5204807 0.5204807
> dim(ovarian)
[1] 26 6
> exp(-basehaz(fit)$hazard)[ovarian$fustat == 1]
[1] 0.9880206 0.9738738 0.9545899 0.8973620 0.8243117 0.8243117 0.7750981
[8] 0.7750981 0.5204807 0.5204807 0.5204807 0.5204807
> summary(survfit(fit))$surv
[1] 0.9880206 0.9738738 0.9545899 0.9334790 0.8973620 0.8624781 0.8243117
[8] 0.7750981 0.7244924 0.6734146 0.5962187 0.5204807
> summary(survfit(fit, data.frame(age=mean(ovarian$age))),
time=ovarian$futime[ovarian$fustat == 1])$surv
# Same result as above
> summary(survfit(fit, data.frame(age=mean(ovarian$age))),
time=ovarian$futime)$surv
[1] 0.9880206 0.9738738 0.9545899 0.9334790 0.8973620 0.8624781 0.8243117
[8] 0.8243117 0.8243117 0.7750981 0.7750981 0.7244924 0.6734146 0.6734146
[15] 0.5962187 0.5204807 0.5204807 0.5204807 0.5204807 0.5204807 0.5204807
[22] 0.5204807 0.5204807 0.5204807 0.5204807 0.5204807
Predicted survival probability in Cox model: survfit.coxph(), plot.survfit() & summary.survfit( , times)
For theory, see section 8.6 Estimation of the survival function in Klein & Moeschberger.
For R, see Extract survival probabilities in Survfit by groups
plot.survfit(). fun="log" to plot log survival curve, fun="event" plot cumulative events, fun="cumhaz" plots cumulative hazard (f(y) = -log(y)).
The plot function below will draw 4 curves: [math]\displaystyle{ S_0(t)^{\exp(\hat{\beta}_{age}*60)} }[/math], [math]\displaystyle{ S_0(t)^{\exp(\hat{\beta}_{age}*60+\hat{\beta}_{stageII})} }[/math], [math]\displaystyle{ S_0(t)^{\exp(\hat{\beta}_{age}*60+\hat{\beta}_{stageIII})} }[/math], [math]\displaystyle{ S_0(t)^{\exp(\hat{\beta}_{age}*60+\hat{\beta}_{stageIV})} }[/math].
library(KMsurv) # Data package for Klein & Moeschberge data(larynx) larynx$stage <- factor(larynx$stage) coxobj <- coxph(Surv(time, delta) ~ age + stage, data = larynx) # Figure 8.3 from Section 8.6 plot(survfit(coxobj, newdata = data.frame(age=rep(60, 4), stage=factor(1:4))), lty = 1:4) # Estimated probability for a 60-year old for different stage patients out <- summary(survfit(coxobj, data.frame(age = rep(60, 4), stage=factor(1:4))), times = 5) out$surv # time n.risk n.event survival1 survival2 survival3 survival4 # 5 34 40 0.702 0.665 0.51 0.142 sum(larynx$time >=5) # n.risk # [1] 34 sum(larynx$delta[larynx$time <=5]) # n.event # [1] 40 sum(larynx$time >5) # Wrong # [1] 31 sum(larynx$delta[larynx$time <5]) # Wrong # [1] 39 # 95% confidence interval out$lower # 0.8629482 0.9102532 0.7352413 0.548579 out$upper # 0.5707952 0.4864903 0.3539527 0.03691768
We need to pay attention when the number of covariates is large (and we don't want to specify each covariates in the formula). The key is to create a data frame and use dot (.) in the formula. This is to fix a warning message: 'newdata' had XXX rows but variables found have YYY rows from running survfit(, newdata).
Another way is to use as.formula() if we don't want to create a new object.
xsub <- data.frame(xtrain)
colnames(xsub) <- paste0("x", 1:ncol(xsub))
coxobj <- coxph(Surv(ytrain[, "time"], ytrain[, "status"]) ~ ., data = xsub)
newdata <- data.frame(xtest)
colnames(newdata) <- paste0("x", 1:ncol(newdata))
survprob <- summary(survfit(coxobj, newdata=newdata),
times = 5)$surv[1, ]
# since there is only 1 time point, we select the first row in surv (surv is a matrix with one row).
Predicted survival probabilities from glmnet: c060/peperr, biospear packages and manual computation
- Terry Therneau: The answer is that you cannot predict survival time, in general
- https://rdrr.io/cran/c060/man/predictProb.glmnet.html
## S3 method for class 'glmnet'
predictProb(object, response, x, times, complexity, ...)
set.seed(1234)
junk <- biospear::simdata(n=500, p=500, q.main = 10, q.inter = 0,
prob.tt = .5, m0=1, alpha.tt=0,
beta.main= -.5, b.corr = .7, b.corr.by=25,
wei.shape = 1, recr=3, fu=2, timefactor=1)
summary(junk$time)
library(glmnet)
library(c060) # Error: object 'predictProb' not found
library(peperr)
y <- cbind(time=junk$time, status=junk$status)
x <- cbind(1, junk[, "treat", drop = FALSE])
names(x) <- c("inter", "treat")
x <- as.matrix(x)
cvfit <- cv.glmnet(x, y, family = "cox")
obj <- glmnet(x, y, family = "cox")
xnew <- matrix(c(0,0), nr=1)
predictProb(obj, y, xnew, times=1, complexity = cvfit$lambda.min)
# Error in exp(lp[response[, 1] >= t.unique[i]]) :
# non-numeric argument to mathematical function
# In addition: Warning message:
# In is.na(x) : is.na() applied to non-(list or vector) of type 'NULL'
- https://www.rdocumentation.org/packages/biospear/versions/1.0.1/topics/expSurv and manual computation (search bhaz)
expSurv(res, traindata, method, ci.level = .95, boot = FALSE, nboot, smooth = TRUE, pct.group = 4, time, trace = TRUE, ncores = 1) # S3 method for resexpSurv predict(object, newdata, ...)
# continue the example
# BMsel() takes a little while
resBM <- biospear::BMsel(
data = junk,
method = "lasso",
inter = FALSE,
folds = 5)
# Note: if we specify time =5 in expsurv(), we will get an error message
# 'time' is out of the range of the observed survival time.
# Note: if we try to specify more than 1 time point, we will get the following msg
# 'time' must be an unique value; no two values are allowed.
esurv <- biospear::expSurv(
res = resBM,
traindata = junk,
boot = FALSE,
time = median(junk$time),
trace = TRUE)
# debug(biospear:::plot.resexpSurv)
plot(esurv, method = "lasso")
# This is equivalent to doing the following
xx <- attributes(esurv)$m.score[, "lasso"]
wc <- order(xx); wgr <- 1:nrow(esurv$surv)
p1 <- plot(x = xx[wc], y = esurv$surv[wgr, "lasso"][wc],
xlab='prognostic score', ylab='survival prob')
# prognostic score beta*x in this cases.
# ignore treatment effect and interactions
xxmy <- as.vector(as.matrix(junk[, names(resBM$lasso)]) %*% resBM$lasso)
# See page4 of the paper. Scaled scores were used in the plot
range(abs(xx - (xxmy-quantile(xxmy, .025)) / (quantile(xxmy, .975) - quantile(xxmy, .025))))
# [1] 1.500431e-09 1.465241e-06
h0 <- bhaz(resBM$lasso, junk$time, junk$status, junk[, names(resBM$lasso)])
newtime <- median(junk$time)
H0 <- sapply(newtime, function(tt) sum(h0$h[h0$dt <= tt]))
# newx <- junk[ , names(resBM$lasso)]
# Compute the estimate of the survival probability at training x and time = median(junk$time)
# using Breslow method
S2 <- outer(exp(-H0), exp(xxmy), "^") # row = newtime, col = newx
range(abs(esurv$surv[wgr, "lasso"] - S2))
# [1] 6.455479e-18 2.459136e-06
# My implementation of the prognostic plot
# Note that the x-axis on the plot is based on prognostic scores beta*x,
# not on treatment modifying scores gamma*x as described in the paper.
# Maybe it is because inter = FALSE in BMsel() we have used.
p2 <- plot(xxmy[wc], S2[wc], xlab='prognostic score', ylab='survival prob') # cf p1
> names(esurv)
[1] "surv" "lower" "upper"
> str(esurv$surv)
num [1:500, 1:2] 0.772 0.886 0.961 0.731 0.749 ...
- attr(*, "dimnames")=List of 2
..$ : NULL
..$ : chr [1:2] "lasso" "oracle"
esurv2 <- predict(esurv, newdata = junk)
esurv2$surv # All zeros?
Bug from the sample data (interaction was considered here; inter = TRUE) ?
set.seed(123456)
resBM <- BMsel(
data = Breast,
x = 4:ncol(Breast),
y = 2:1,
tt = 3,
inter = TRUE,
std.x = TRUE,
folds = 5,
method = c("lasso", "lasso-pcvl"))
esurv <- expSurv(
res = resBM,
traindata = Breast,
boot = FALSE,
smooth = TRUE,
time = 4,
trace = TRUE
)
Computation of the expected survival
Computation of analytical confidence intervals
Computation of smoothed B-splines
Error in cobs(x = x, y = y, print.mesg = F, print.warn = F, method = "uniform", :
There is at least one pair of adjacent knots that contains no observation.
Loglikelihood
- fit$loglik is a vector of length 2 (Null model, fitted model)
- Use survival::anova() command to do a likelihood ratio test. Note this function does not work on glmnet object.
- residuals.coxph Calculates martingale, deviance, score or Schoenfeld residuals for a Cox proportional hazards model.
- No deviance() on coxph object!
- logLik() function will return fit$loglik[2]
glmnet
- It seems AIC does not require the assumption of nested models.
- https://en.wikipedia.org/wiki/Akaike_information_criterion, (akaike pronunciation in Japanese)
- [math]\displaystyle{ \begin{align} \mathrm{AIC} &= 2k - 2\ln(\hat L) \\ \mathrm{AICc} &= \mathrm{AIC} + \frac{2k^2 + 2k}{n - k - 1} \end{align} }[/math]
- Is it possible to calculate AIC and BIC for lasso regression models?. See the references about the degrees of freedom in Lasso regressions.
fit <- glmnet(x, y, family = "multinomial") tLL <- fit$nulldev - deviance(fit) # ln(L) k <- fit$df n <- fit$nobs AICc <- -tLL+2*k+2*k*(k+1)/(n-k-1) AICc
- For glmnet object, see ?deviance.glmnet and R: Getting AIC/BIC/Likelihood from GLMNet. An example with all continuous variables
set.seed(10101) N=1000;p=6 nzc=p/3 x=matrix(rnorm(N*p),N,p) beta=rnorm(nzc) fx=x[,seq(nzc)]%*%beta/3 hx=exp(fx) ty=rexp(N,hx) tcens=rbinom(n=N,prob=.3,size=1)# censoring indicator y=cbind(time=ty,status=1-tcens) # y=Surv(ty,1-tcens) with library(survival) coxobj <- coxph(Surv(ty, 1-tcens) ~ x) coxobj_small <- coxph(Surv(ty, 1-tcens) ~1) anova(coxobj, coxobj_small) # Analysis of Deviance Table # Cox model: response is Surv(ty, 1 - tcens) # Model 1: ~ x # Model 2: ~ 1 # loglik Chisq Df P(>|Chi|) # 1 -4095.2 # 2 -4102.7 15.151 6 0.01911 * fit2 <- glmnet(x,y,family="cox", lambda=0) # ridge regression deviance(fit2) # [1] 8190.313 fit2$df # [1] 6 fit2$nulldev - deviance(fit2) # Log-Likelihood ratio statistic # [1] 15.15097 1-pchisq(fit2$nulldev - deviance(fit2), fit2$df) # [1] 0.01911446
Here is another example including a factor covariate:
library(KMsurv) # Data package for Klein & Moeschberge data(larynx) larynx$stage <- factor(larynx$stage) coxobj <- coxph(Surv(time, delta) ~ age + stage, data = larynx) coef(coxobj) # age stage2 stage3 stage4 # 0.0190311 0.1400402 0.6423817 1.7059796 coxobj_small <- coxph(Surv(time, delta)~age, data = larynx) anova(coxobj, coxobj_small) # Analysis of Deviance Table # Cox model: response is Surv(time, delta) # Model 1: ~ age + stage # Model 2: ~ age # loglik Chisq Df P(>|Chi|) # 1 -187.71 # 2 -195.55 15.681 3 0.001318 ** # --- # Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 # Now let's look at the glmnet() function. # It seems glmnet does not recognize factor covariates. coxobj2 <- with(larynx, glmnet(cbind(age, stage), Surv(time, delta), family = "cox", lambda=0)) coxobj2$nulldev - deviance(coxobj2) # Log-Likelihood ratio statistic # [1] 15.72596 coxobj1 <- with(larynx, glmnet(cbind(1, age), Surv(time, delta), family = "cox", lambda=0)) deviance(coxobj1) - deviance(coxobj2) # [1] 13.08457 1-pchisq(deviance(coxobj1) - deviance(coxobj2) , coxobj2$df-coxobj1$df) # [1] 0.0002977376
High dimensional data
https://cran.r-project.org/web/views/Survival.html
glmnet + Cox models
- Robust estimation of the expected survival probabilities from high-dimensional Cox models with biomarker-by-treatment interactions in randomized clinical trials by Nils Ternès, Federico Rotolo and Stefan Michiels, BMC Medical Research Methodology, 2017 (open review available). The corresponding software biospear on cran and rdocumentation.org.
- Expected time of survival in glmnet for cox family
Error in glmnet: x should be a matrix with 2 or more columns
Error in coxnet: (list) object cannot be coerced to type 'double'
Fix: do not use data.frame in X. Use cbind() instead.
Prognostic index/risk scores
- International Prognostic Index
- The test data were first segregated into high-risk and low-risk groups by the median of training risk scores. Assessment of performance of survival prediction models for cancer prognosis
linear.predictors component in coxph object
The $linear.predictors component is not [math]\displaystyle{ \beta' x }[/math]. It is [math]\displaystyle{ \beta' (x-\mu_x) }[/math]. See this post.
predict.coxph, prognostic index & risk score
- predict.coxph() Compute fitted values and regression terms for a model fitted by coxph. The Cox model is a relative risk model; predictions of type "linear predictor", "risk", and "terms" are all relative to the sample from which they came. By default, the reference value for each of these is the mean covariate within strata. The primary underlying reason is statistical: a Cox model only predicts relative risks between pairs of subjects within the same strata, and hence the addition of a constant to any covariate, either overall or only within a particular stratum, has no effect on the fitted results. Returned value: a vector or matrix of predictions, or a list containing the predictions (element "fit") and their standard errors (element "se.fit") if the se.fit option is TRUE.
predict(object, newdata,
type=c("lp", "risk", "expected", "terms", "survival"),
se.fit=FALSE, na.action=na.pass, terms=names(object$assign), collapse,
reference=c("strata", "sample"), ...)
- http://stats.stackexchange.com/questions/44896/how-to-interpret-the-output-of-predict-coxph. The $linear.predictors component represents [math]\displaystyle{ \beta (x - \bar{x}) }[/math]. The risk score (type='risk') corresponds to [math]\displaystyle{ \exp(\beta (x-\bar{x})) }[/math]. Factors are converted to dummy predictors as usual; see model.matrix.
- http://www.togaware.com/datamining/survivor/Lung1.html
library(coxph) fit <- coxph(Surv(time, status) ~ age , lung) fit # Call: # coxph(formula = Surv(time, status) ~ age, data = lung) # coef exp(coef) se(coef) z p # age 0.0187 1.02 0.0092 2.03 0.042 # # Likelihood ratio test=4.24 on 1 df, p=0.0395 n= 228, number of events= 165 fit$means # age # 62.44737 # type = "lr" (Linear predictor) as.numeric(predict(fit,type="lp"))[1:10] # [1] 0.21626733 0.10394626 -0.12069589 -0.10197571 -0.04581518 0.21626733 # [7] 0.10394626 0.16010680 -0.17685643 -0.02709500 0.0187 * (lung$age[1:10] - fit$means) # [1] 0.21603421 0.10383421 -0.12056579 -0.10186579 -0.04576579 0.21603421 # [7] 0.10383421 0.15993421 -0.17666579 -0.02706579 fit$linear.predictors[1:10] # [1] 0.21626733 0.10394626 -0.12069589 -0.10197571 -0.04581518 # [6] 0.21626733 0.10394626 0.16010680 -0.17685643 -0.02709500 # type = "risk" (Risk score) > as.numeric(predict(fit,type="risk"))[1:10] [1] 1.2414342 1.1095408 0.8863035 0.9030515 0.9552185 1.2414342 1.1095408 [8] 1.1736362 0.8379001 0.9732688 > exp((lung$age-mean(lung$age)) * 0.0187)[1:10] [1] 1.2411448 1.1094165 0.8864188 0.9031508 0.9552657 1.2411448 [7] 1.1094165 1.1734337 0.8380598 0.9732972 > exp(fit$linear.predictors)[1:10] [1] 1.2414342 1.1095408 0.8863035 0.9030515 0.9552185 1.2414342 [7] 1.1095408 1.1736362 0.8379001 0.9732688
Survival risk prediction
- Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data Simon 2011. The authors have noted the CV has been used for optimization of tuning parameters but the data available are too limited for effective into training & test sets.
- The CV Kaplan-Meier curves are essentially unbiased and the separation between the curves gives a fair representation of the value of the expression profiles for predicting survival risk.
- The log-rank statistic does not have the usual chi-squared distribution under the null hypothesis. This is because the data was used to create the risk groups.
- Survival ROC curve can be used as a measure of predictive accuracy for the survival risk group model at a certain landmark time.
- The ROC curve can be misleading. For example if re-substitution is used, the AUC can be very large.
- The p-value for the significance of the test that AUC=.5 for the cross-validated survival ROC curve can be computed by permutations.
- An evaluation of resampling methods for assessment of survival risk prediction in high-dimensional settings Subramanian, et al 2010.
- Measure of assessment for prognostic prediction [math]\displaystyle{ \mbox{Sensitivity}(c,t) = P(\beta' X \gt c | T \lt t), \mbox{Specificity}(c, t) = P(\beta' X \le c | T \ge t) }[/math] Recall that in the categorical response data [math]\displaystyle{ \mbox{Sensitivity}=P(Pred=1|True=1), \mbox{Specificity}=P(Pred=0|True=0) }[/math]
- The conditional probabilities can be estimated by Heagerty et al 2000 (R package 'survivalROC'). The AUC(t) can be used for comparing and assessing prognostic models (a measure of accuracy) for future samples. In the absence of an independent large dataset, an estimate for AUC(t) is obtained through resampling from the original sample [math]\displaystyle{ S_n }[/math].
- Resubstitution estimate of AUC(t) (i.e. all observations were used for feature selection, model building as well as the estimation of accuracy) is too optimistic. So k-fold CV method is considered.
- There are two ways to compute k-fold CV estimate of AUC(t): the pooling strategy (used in the paper) and average strategy (AUC(t)s are first computed for each test set and are then averaged). In the pooling strategy, all the test set risk-score predictions are first collected and AUC(t) is calculated on this combined set.
- Conclusions: sample splitting and LOOCV have a higher mean square error than other methods. 5-fold or 10-fold CV provide a good balance between bias and variability for a wide range of data settings.
- Gene Expression–Based Prognostic Signatures in Lung Cancer: Ready for Clinical Use? Subramanian, et al 2010.
- Assessment of survival prediction models based on microarray data Martin Schumacher, et al. 2007
- Semi-Supervised Methods to Predict Patient Survival from Gene Expression Data Eric Bair , Robert Tibshirani, 2004
- Time dependent ROC curves for censored survival data and a diagnostic marker. Heagerty et al, Biometrics 2000
- An introduction to survivalROC by Saha, Heagerty. If the AUCs are computed at several time points, we can plot the AUCs vs time for different models (eg different covariates) and compare them to see which model performs better.
- Assessment of Discrimination in Survival Analysis (C-statistics, etc)
- Survival prediction from clinico-genomic models - a comparative study Hege M Bøvelstad, 2009
- Assessment and comparison of prognostic classification schemes for survival data. E. Graf, C. Schmoor, W. Sauerbrei, et al. 1999
- What do we mean by validating a prognostic model? Douglas G. Altman, Patrick Royston, 2000
- On the prognostic value of survival models with application to gene expression signatures T. Hielscher, M. Zucknick, W. Werft, A. Benner, 2000
- Assessment of performance of survival prediction models for cancer prognosis Hung-Chia Chen et al 2012
- Accuracy of predictive ability measures for survival models Flandre, Detsch and O'Quigley, 2017.
- Association between expression of random gene sets and survival is evident in multiple cancer types and may be explained by sub-classification Yishai Shimoni, PLOS 2018
- Development and validation of risk prediction equations to estimate survival in patients with colorectal cancer: cohort study
- Cox-nnet: An artificial neural network method for prognosis prediction of high-throughput omics data Ching et al 2018.
Assessing the performance of prediction models
- Investigating the prediction ability of survival models based on both clinical and omics data: two case studies by Riccardo De Bin, Statistics in Medicine 2014. (not useful)
- Assessment of performance of survival prediction models for cancer prognosis Chen et al , BMC Medical Research Methodology 2012
- A simulation study of predictive ability measures in a survival model I: Explained variation measures Choodari‐Oskooei et al, Stat in Medicine 2011
- Assessing the performance of prediction models: a framework for some traditional and novel measures by Ewout W. Steyerberg, Andrew J. Vickers, [...], and Michael W. Kattan, 2010.
- survcomp: an R/Bioconductor package for performance assessment and comparison of survival models paper in 2011 and Introduction to R and Bioconductor Survival analysis where the survcomp package can be used. The summary here is based on this paper.
- How to compare predictive power of survival models?
- How to compare Harrell C-index from different models in survival analysis? and Frank Harrell's comment: Likelihood ratio test give you a powerful test.
Hazard ratio
hazard.ratio(x, surv.time, surv.event, weights, strat, alpha = 0.05,
method.test = c("logrank", "likelihood.ratio", "wald"), na.rm = FALSE, ...)
D index
D.index(x, surv.time, surv.event, weights, strat, alpha = 0.05,
method.test = c("logrank", "likelihood.ratio", "wald"), na.rm = FALSE, ...)
Concordance index (C-index)
- The area under ROC curve (plot of sensitivity of 1-specificity) is also called C-statistic. It is a measure of discrimination generalized for survival data (Harrell 1982 & 2001). The ROC are functions of the sensitivity and specificity for each value of the measure of model. (Nancy Cook, 2007)
- The sensitivity of a test is the probability of a positive test result, or of a value above a threshold, among those with disease (cases).
- The specificity of a test is the probability of a negative test result, or of a value below a threshold, among those without disease (noncases).
- Perfect discrimination corresponds to a c-statistic of 1 & is achieved if the scores for all the cases are higher than those for all the non-cases.
- The c-statistic is the probability that the measure or predicted risk is higher for a case than for a noncase.
- The c-statistic is not the probability that individuals are classified correctly or that a person with a high test score will eventually become a case.
- C-statistic is a rank-based measure. The c-statistic describes how well models can rank order cases and noncases, but not a function of the actual predicted probabilities.
- How to interpret the output for calculating concordance index (c-index)? [math]\displaystyle{
P(\beta' Z_1 \gt \beta' Z_2|T_1 \lt T_2)
}[/math] where T is the survival time and Z is the covariates.
- It is the fraction of pairs in your data, where the observation with the higher survival time has the higher probability of survival predicted by your model.
- High values mean that your model predicts higher probabilities of survival for higher observed survival times.
- The c index estimates the probability of concordance between predicted and observed responses. A value of 0.5 indicates no predictive discrimination and a value of 1.0 indicates perfect separation of patients with different outcomes. (p371 Harrell 1996)
- Drawback of C-statistics:
- Even though rank indexes such as c are widely applicable and easily interpretable, they are not sensitive for detecting small differences in discrimination ability between two models. This is due to the fact that a rank method considers the (prediction, outcome) pairs (0.01,0), (0.9, 1) as no more concordant than the pairs (0.05,0), (0.8, 1). A more sensitive likelihood-ratio Chi-square-based statistic that reduces to R2 in the linear regression case may be substituted. (p371 Harrell 1996)
- If the model is correct, the likelihood based measures may be more sensitive in detecting differences in prediction ability, compared to rank-based measures such as C-indexes. (Uno 2011 p 1113)
- http://dmkd.cs.vt.edu/TUTORIAL/Survival/Slides.pdf
- survcomp package
- concordance.index()
- Hmisc package. rcorr.cens().
See also 5 Ways to Estimate Concordance Index for Cox Models in R, Why Results Aren't Identical?
Integrated brier score
Assessment and comparison of prognostic classification schemes for survival data Graf et al Stat. Med. 1999 2529-45
- Because the point predictions of event-free times will almost inevitably given inaccurate and unsatisfactory result, the mean square error of prediction [math]\displaystyle{ \frac{1}{n}\sum_1^n (T_i - \hat{T}(X_i))^2 }[/math] method will not be considered.
- Another approach is to predict the survival or event status [math]\displaystyle{ Y=I(T \gt \tau) }[/math] at a fixed time point [math]\displaystyle{ \tau }[/math] for a patient with X=x. This leads to the expected Brier score [math]\displaystyle{ E[(Y - \hat{S}(\tau|X))^2] }[/math] where [math]\displaystyle{ \hat{S}(\tau|X) }[/math] is the estimated event-free probabilities (survival probability) at time [math]\displaystyle{ \tau }[/math] for subject with predictor variable [math]\displaystyle{ X }[/math].
- The time-dependent Brier score (without censoring)
- [math]\displaystyle{ \begin{align} \mbox{Brier}(\tau) &= \frac{1}{n}\sum_1^n (I(T_i\gt \tau) - \hat{S}(\tau|X_i))^2 \end{align} }[/math]
- The time-dependent Brier score (with censoring, C is the censoring variable)
- [math]\displaystyle{ \begin{align} \mbox{Brier}(\tau) = \frac{1}{n}\sum_i^n\bigg[\frac{(\hat{S}_C(t_i))^2I(t_i \leq \tau, \delta_i=1)}{\hat{S}_C(t_i)} + \frac{(1 - \hat{S}_C(t_i))^2 I(t_i \gt \tau)}{\hat{S}_C(\tau)}\bigg] \end{align} }[/math]
where [math]\displaystyle{ \hat{S}_C(t_i) = P(C \gt t_i) }[/math], the Kaplan-Meier estimate of the censoring distribution with [math]\displaystyle{ t_i }[/math] the survival time of patient i. The integration of the Brier score can be done by over time [math]\displaystyle{ t \in [0, \tau] }[/math] with respect to some weight function W(t) for which a natual choice is [math]\displaystyle{ (1 - \hat{S}(t))/(1-\hat{S}(\tau)) }[/math]. The lower the iBrier score, the larger the prediction accuracy is.
- Useful benchmark values for the Brier score are 33%, which corresponds to predicting the risk by a random number drawn from U[0, 1], and 25% which corresponds to predicting 50% risk for everyone. See Evaluating Random Forests for Survival Analysis using Prediction Error Curves by Mogensen et al J. Stat Software 2012 (pec package). The paper has a good summary of different R package implementing Brier scores.
R function
- pec by Thomas A. Gerds
- peperr package. The package peperr is an early branch of pec.
- survcomp::sbrier.score2proba().
- ipred::sbrier()
Papers on high dimensional covariates
- Assessment of survival prediction models based on microarray data, Bioinformatics , 2007, vol. 23 (pg. 1768-74)
- Allowing for mandatory covariates in boosting estimation of sparse high-dimensional survival models, BMC Bioinformatics , 2008, vol. 9 pg. 14
C-statistics
- C-statistics is the probability of concordance between predicted and observed survival.
- Harrell’s Concordance. In SAS, the s.e. of the Harrell's C-statistics is estimated by the delta method as described in Comparing two correlated C indices with right‐censored survival outcome: a one‐shot nonparametric approach. [math]\displaystyle{ \begin{align} C_H = \frac{\sum_{i,j}I(t_i \lt t_{j}) I(\hat{\beta} Z_i \gt \hat{\beta} Z_j) \delta_i}{\sum_{i,j} I(t_i \lt t_j) \delta_i} \end{align} }[/math] which converges to a censoring-dependent quantity [math]\displaystyle{ P(\beta'Z_1 \gt \beta' Z_2|T_1 \lt T_2, T_1 \lt \text{min}(D_1,D_2)). }[/math] Here D is the censoring variable.
- On the C-statistics for Evaluating Overall Adequacy of Risk Prediction Procedures with Censored Survival Data by Uno et al 2011. Let [math]\displaystyle{ \tau }[/math] be a specified time point within the support of the censoring variable. [math]\displaystyle{
\begin{align}
C(\tau) = \text{UnoC}(\hat{\pi}, \tau) = \frac{\sum_{i,i'}(\hat{S}_C(t_i))^{-2}I(t_i \lt t_{i'}, t_i \lt \tau) I(\hat{\pi}_i \gt \hat{\pi}_{i'}) \delta_i}{\sum_{i,i'}(\hat{S}_C(t_i))^{-2}I(t_i \lt t_{i'}, t_i \lt \tau) \delta_i}
\end{align}
}[/math], a measure of the concordance between [math]\displaystyle{ \hat{\pi}_i = \hat{\beta} Z_i }[/math] (the linear predictor) and the survival time. [math]\displaystyle{ \hat{S}_C(t) }[/math] is the Kaplan-Meier estimator for the censoring distribution/variable/time; flipping the definition of [math]\displaystyle{ \delta_i }[/math]/considering failure events as "censored" observations and censored observations as "failures" and computing the KM as usual; see p207 of Satten 2001 and the source code from the kmcens() in survC1. Note that [math]\displaystyle{ C_\tau }[/math] converges to [math]\displaystyle{ P(\beta'Z_1 \gt \beta' Z_2|T_1 \lt T_2, T_1 \lt \tau). }[/math]
- real data 1: fit a Cox model. Get risk scores [math]\displaystyle{ \hat{\beta}'Z }[/math]. Compute the point and confidence interval estimates (M=500 indep. random samples with the same sample size as the observation data) of [math]\displaystyle{ C_\tau }[/math] for different [math]\displaystyle{ \tau }[/math]. Compare them with the conventional C-index procedure (Korn).
- real data 1: compute [math]\displaystyle{ C_\tau }[/math] for a full model and a reduce model. Compute the difference of them ([math]\displaystyle{ C_\tau^{(A)} - C_\tau^{(B)} = .01 }[/math]) and the 95% confidence interval (-0.00, .02) of the difference for testing the importance of some variable (HDL in this case). Though HDL is quite significant (p=0) with respect to the risk of CV disease but its incremental value evaluated via C-statistics is quite modest.
- real data 2: goal - evaluate the prognostic value of a new gene signature in predicting the time to death or metastasis for breast cancer patients. Two models were fitted; one with age+ER and the other is gene+age+ER. For each model we can calculate the point and interval estimates of [math]\displaystyle{ C_\tau }[/math] for different [math]\displaystyle{ \tau }[/math]s.
- simulation: T is from Weibull regression for case 1 and log-normal regression for case 2. Covariates = (age, ER, gene). 3 kinds of censoring were considered. Sample size is 100, 150, 200 and 300. 1000 iterations. Compute coverage probabilities and average length of 95% confidence intervals, bias and root mean square error for [math]\displaystyle{ \tau }[/math] equals to 10 and 15. Compared with the conventional approach, the new method has higher coverage probabilities and less bias in 6 scenarios.
- Statistical methods for the assessment of prognostic biomarkers (Part I): Discrimination by Tripep et al 2010
- Assessment of Discrimination in Survival Analysis (C-statistics, etc) by anonymous
- Uno's C-statistics
- It seems C-statistics is a decreasing function of tau.
- (From ?UnoC) Uno's estimator is based on inverse-probability-of-censoring weights and does not assume a specific working model for deriving the predictor lpnew. It is assumed, however, that there is a one-to-one relationship between the predictor and the expected survival times conditional on the predictor. Note that the estimator implemented in UnoC is restricted to situations where the random censoring assumption holds.
- UnoC() from the survAUC package. It can take new data. The tau parameter: Truncation time. The resulting C tells how well the given prediction model works in predicting events that occur in the time range from 0 to tau. Con: no confidence interval estimate for [math]\displaystyle{ C_\tau }[/math] nor [math]\displaystyle{ C_\tau^{(A)} - C_\tau^{(B)} }[/math].
- Est.Cval() from the survC1 package (authored by H. Uno). It doesn't take new data nor the vector of predictors obtained from the test data. Pro: Inf.Cval() can compute the CI of [math]\displaystyle{ C_\tau }[/math] & Inf.Cval.Delta() for the difference [math]\displaystyle{ C_\tau^{(A)} - C_\tau^{(B)} }[/math].
library(survAUC)
# require training and predict sets
TR <- ovarian[1:16,]
TE <- ovarian[17:26,]
train.fit <- coxph(Surv(futime, fustat) ~ age, data=TR)
lpnew <- predict(train.fit, newdata=TE)
Surv.rsp <- Surv(TR$futime, TR$fustat)
Surv.rsp.new <- Surv(TE$futime, TE$fustat)
UnoC(Surv.rsp, Surv.rsp, train.fit$linear.predictors, time=365.25*1)
# [1] 0.9761905
UnoC(Surv.rsp, Surv.rsp, train.fit$linear.predictors, time=365.25*2)
# [1] 0.7308979
UnoC(Surv.rsp, Surv.rsp, train.fit$linear.predictors, time=365.25*3)
# [1] 0.7308979
UnoC(Surv.rsp, Surv.rsp, train.fit$linear.predictors, time=365.25*4)
# [1] 0.7308979
UnoC(Surv.rsp, Surv.rsp, train.fit$linear.predictors, time=365.25*5)
# [1] 0.7308979
UnoC(Surv.rsp, Surv.rsp, train.fit$linear.predictors)
# [1] 0.7308979
# So the function UnoC() can obtain the exact result as Est.Cval().
# Now try on a new data set. Question: why do we need Surv.rsp?
UnoC(Surv.rsp, Surv.rsp.new, lpnew)
# [1] 0.7333333
UnoC(Surv.rsp, Surv.rsp.new, lpnew, time=365.25*2)
# [1] 0.7333333
library(survC1)
# tau is mandatory (>0), no need to have training and predict sets
Est.Cval(ovarian[1:16, c(1,2, 3)], tau=365.25*1)$Dhat
# [1] 0.9761905
Est.Cval(ovarian[1:16, c(1,2, 3)], tau=365.25*2)$Dhat
# [1] 0.7308979
Est.Cval(ovarian[1:16, c(1,2, 3)], tau=365.25*3)$Dhat
# [1] 0.7308979
Est.Cval(ovarian[1:16, c(1,2, 3)], tau=365.25*4)$Dhat
# [1] 0.7308979
Est.Cval(ovarian[1:16, c(1,2, 3)], tau=365.25*5)$Dhat
# [1] 0.7308979
svg("~/Downloads/c_stat_scatter.svg", width=8, height=5)
par(mfrow=c(1,2))
plot(TR$futime, train.fit$linear.predictors, main="training data", xlab="time", ylab="predictor")
mtext("C=.731 at t=2", 3)
plot(TE$futime, lpnew, main="testing data", xlab="time", ylab="predictor")
mtext("C=.733 at t=2", 3)
dev.off()
- 5 Ways to Estimate Concordance Index for Cox Models in R, Why Results Aren't Identical?, C-index/C-statistic 计算的5种不同方法及比较. The 5 functions are rcorrcens() from Hmisc, summary()$concordance from survival, survConcordance() from survival, concordance.index() from survcomp and cph() from rms.
- Assessing the prediction accuracy of a cure model for censored survival data with long-term survivors: Application to breast cancer data
- The use of ROC for defining the validity of the prognostic index in censored data
- Use and Misuse of the Receiver Operating Characteristic Curve in Risk Prediction Cook 2007
- The c-index is not proper for the evaluation of t-year predicted risks by Paul Blanche, 2018
C-statistic vs LRT comparing nested models
1. Binary data
# https://stats.stackexchange.com/questions/46523/how-to-simulate-artificial-data-for-logistic-regression
set.seed(666)
x1 = rnorm(1000) # some continuous variables
x2 = rnorm(1000)
z = 1 + 2*x1 + 3*x2 # linear combination with a bias
pr = 1/(1+exp(-z)) # pass through an inv-logit function
y = rbinom(1000,1,pr) # bernoulli response variable
df = data.frame(y=y,x1=x1,x2=x2)
fit <- glm( y~x1+x2,data=df,family="binomial")
summary(fit)
# Estimate Std. Error z value Pr(>|z|)
# (Intercept) 0.9915 0.1185 8.367 <2e-16 ***
# x1 2.2731 0.1789 12.709 <2e-16 ***
# x2 3.1853 0.2157 14.768 <2e-16 ***
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#
# (Dispersion parameter for binomial family taken to be 1)
#
# Null deviance: 1355.16 on 999 degrees of freedom
# Residual deviance: 582.93 on 997 degrees of freedom
# AIC: 588.93
confint.default(fit)
# 2.5 % 97.5 %
# (Intercept) 0.7592637 1.223790
# x1 1.9225261 2.623659
# x2 2.7625861 3.608069
# LRT - likelihood ratio test
fit2 <- glm( y~x1,data=df,family="binomial")
anova.res <- anova(fit2, fit)
# Analysis of Deviance Table
#
# Model 1: y ~ x1
# Model 2: y ~ x1 + x2
# Resid. Df Resid. Dev Df Deviance
# 1 998 1186.16
# 2 997 582.93 1 603.23
1-pchisq( abs(anova.res$Deviance[2]), abs(anova.res$Df[2]))
# [1] 0
# Method 1: use ROC package to compute AUC
library(ROC)
set.seed(123)
markers <- predict(fit, newdata = data.frame(x1, x2), type = "response")
roc1 <- rocdemo.sca( truth=y, data=markers, rule=dxrule.sca )
auc <- AUC(roc1); print(auc) # [1] 0.9459085
markers2 <- predict(fit2, newdata = data.frame(x1), type = "response")
roc2 <- rocdemo.sca( truth=y, data=markers2, rule=dxrule.sca )
auc2 <- AUC(roc2); print(auc2) # [1] 0.7259098
auc - auc2 # [1] 0.2199987
# Method 2: use pROC package to compute AUC
roc_obj <- pROC::roc(y, markers)
pROC::auc(roc_obj) # Area under the curve: 0.9459
# Method 3: Compute AUC by hand
# https://www.r-bloggers.com/calculating-auc-the-area-under-a-roc-curve/
auc_probability <- function(labels, scores, N=1e7){
pos <- sample(scores[labels], N, replace=TRUE)
neg <- sample(scores[!labels], N, replace=TRUE)
# sum( (1 + sign(pos - neg))/2)/N # does the same thing
(sum(pos > neg) + sum(pos == neg)/2) / N # give partial credit for ties
}
auc_probability(as.logical(y), markers) # [1] 0.945964
2. Survival data
library(survival)
data(ovarian)
head(ovarian)
range(ovarian$futime) # [1] 59 1227
plot(survfit(Surv(futime, fustat) ~ 1, data = ovarian))
coxph(Surv(futime, fustat) ~ rx + age, data = ovarian)
# coef exp(coef) se(coef) z p
# rx -0.8040 0.4475 0.6320 -1.27 0.2034
# age 0.1473 1.1587 0.0461 3.19 0.0014
#
# Likelihood ratio test=15.9 on 2 df, p=0.000355
# n= 26, number of events= 12
require(survC1)
covs0 <- as.matrix(ovarian[, c("rx")])
covs1 <- as.matrix(ovarian[, c("rx", "age")])
tau=365.25*1
Delta=Inf.Cval.Delta(ovarian[, 1:2], covs0, covs1, tau, itr=200)
round(Delta, digits=3)
# Est SE Lower95 Upper95
# Model1 0.844 0.119 0.611 1.077
# Model0 0.659 0.148 0.369 0.949
# Delta 0.185 0.197 -0.201 0.572
Time dependent ROC curves
Prognostic markers vs predictive markers
- Prognostic marker inform about likely disease outcome independent of the treatment received. See Statistical and practical considerations for clinical evaluation of predictive biomarkers by Mei-Yin Polley et al 2013.
- Predictive marker/treatment selection markers provide information about likely outcomes with application of specific interventions. See Measuring the performance of markers for guiding treatment decisions by Janes, et al 2011.
- Statistical controversies in clinical research: prognostic gene signatures are not (yet) useful in clinical practice by Michiels 2016.
Computation for gene expression (microarray) data
- survival package (basic package, not designed for gene expression)
- gsa package
- samr package
- pamr package
- (Bioconductor) genefilter, source. genefilter() & coxfilter(). apply() was used.
- logpl() from survcomp package
n <- 500
g <- 10000
y <- rexp(n)
status <- ifelse(runif(n) < .7, 1, 0)
x <- matrix(rnorm(n*g), nr=g)
treat <- rbinom(n, 1, .5)
# Method 1
system.time(for(i in 1:g) coxph(Surv(y, status) ~ x[i, ] + treat + treat:x[i, ]))
# 28 seconds
# Method 2
system.time(apply(x, 1, function(z) coxph(Surv(y, status) ~ z + treat + treat:z)))
# 29 seconds
# Method 3 (Windows)
dyn.load("C:/Program Files (x86)/ArrayTools/Fortran/surv64.dll")
tme <- y
sorted <- order(tme)
stime <- as.double(tme[sorted])
sstat <- as.integer(status[sorted])
x1 <- x[,sorted]
imodel <- 1 # imodel=1, fit univariate gene expression. Return p-values vector.
nvar <- 1
system.time(outx1 <- .Fortran("coxfitc", as.integer(n), as.integer(g), as.integer(0),
stime, sstat, t(x1), as.double(0), as.integer(imodel),
double(2*n+2*nvar*nvar+3*nvar), logdiff = double(g)))
# 1.69 seconds on R i386
# 0.79 seconds on R x64
# method 4: GSA
genenames=paste("g", 1:g, sep="")
#create some random gene sets
genesets=vector("list", 50)
for(i in 1:50){
genesets[[i]]=paste("g", sample(1:g,size=30), sep="")
}
geneset.names=paste("set",as.character(1:50),sep="")
debug(GSA.func)
GSA.obj<-GSA(x,y, genenames=genenames, genesets=genesets,
censoring.status=status,
resp.type="Survival", nperms=1)
Browse[3]> str(catalog.unique)
int [1:1401] 7943 227 4069 3011 8402 1586 2443 2777 673 9021 ...
Browse[3]> system.time(cox.func(x[catalog.unique,], y, censoring.status, s0=0))
# 1.3 seconds
Browse[2]> system.time(cox.func(x, y, censoring.status, s0=0))
# 7.259 seconds
More
- This pdf file from data.princeton.edu contains estimation, hypothesis testing, time varying covariates and baseline survival estimation.
- Survival analysis: basic terms, the exponential model, censoring, examples in R and JAGS
- Survival analysis is not commonly used to predict future times to an event. Cox model would require specification of the baseline hazard function.
Logistic regression
Simulate binary data from the logistic model
set.seed(666) x1 = rnorm(1000) # some continuous variables x2 = rnorm(1000) z = 1 + 2*x1 + 3*x2 # linear combination with a bias pr = 1/(1+exp(-z)) # pass through an inv-logit function y = rbinom(1000,1,pr) # bernoulli response variable #now feed it to glm: df = data.frame(y=y,x1=x1,x2=x2) glm( y~x1+x2,data=df,family="binomial")
Building a Logistic Regression model from scratch
https://www.analyticsvidhya.com/blog/2015/10/basics-logistic-regression
Odds ratio
Calculate the odds ratio from the coefficient estimates; see this post.
require(MASS)
N <- 100 # generate some data
X1 <- rnorm(N, 175, 7)
X2 <- rnorm(N, 30, 8)
X3 <- abs(rnorm(N, 60, 30))
Y <- 0.5*X1 - 0.3*X2 - 0.4*X3 + 10 + rnorm(N, 0, 12)
# dichotomize Y and do logistic regression
Yfac <- cut(Y, breaks=c(-Inf, median(Y), Inf), labels=c("lo", "hi"))
glmFit <- glm(Yfac ~ X1 + X2 + X3, family=binomial(link="logit"))
exp(cbind(coef(glmFit), confint(glmFit)))
Medical applications
Subgroup analysis
Other related keywords: recursive partitioning, randomized clinical trials (RCT)
- Thinking about different ways to analyze sub-groups in an RCT
- Tutorial in biostatistics: data-driven subgroup identification and analysis in clinical trials I Lipkovich, A Dmitrienko - Statistics in medicine, 2017
- Personalized medicine:Four perspectives of tailored medicine SJ Ruberg, L Shen - Statistics in Biopharmaceutical Research, 2015
- Berger, J. O., Wang, X., and Shen, L. (2014), “A Bayesian Approach to Subgroup Identification,” Journal of Biopharmaceutical Statistics, 24, 110–129.
Interaction analysis
- Goal: assessing the predictiveness of biomarkers by testing their interaction (strength) with the treatment.
- http://www.stat.purdue.edu/~ghobbs/STAT_512/Lecture_Notes/ANOVA/Topic_27.pdf#page=15. For survival data, y-axis is the survival time and B1=treatment, B2=control and X-axis is treatment-effect modifying score. But as seen on page16, the effects may not be separated.
- Identification of biomarker-by-treatment interactions in randomized clinical trials with survival outcomes and high-dimensional spaces N Ternès, F Rotolo, G Heinze, S Michiels - Biometrical Journal, 2017
- Tian, L., Alizaden, A. A., Gentles, A. J., and Tibshirani, R. (2014) “A Simple Method for Detecting Interactions Between a Treatment and a Large Number of Covariates,” and the book chapter.
- Gunter, L., Zhu, J., and Murphy, S. (2011), “Variable Selection for Qualitative Interactions in Personalized Medicine While Controlling the Family-Wise Error Rate,” Journal of Biopharmaceutical Statistics, 21, 1063–1078.
Statistical Learning
- Elements of Statistical Learning Book homepage
- From Linear Models to Machine Learning by Norman Matloff
- 10 Free Must-Read Books for Machine Learning and Data Science
- 10 Statistical Techniques Data Scientists Need to Master
- Linear regression
- Classification: Logistic Regression, Linear Discriminant Analysis, Quadratic Discriminant Analysis
- Resampling methods: Bootstrapping and Cross-Validation
- Subset selection: Best-Subset Selection, Forward Stepwise Selection, Backward Stepwise Selection, Hybrid Methods
- Shrinkage/regularization: Ridge regression, Lasso
- Dimension reduction: Principal Components Regression, Partial least squares
- Nonlinear models: Piecewise function, Spline, generalized additive model
- Tree-based methods: Bagging, Boosting, Random Forest
- Support vector machine
- Unsupervised learning: PCA, k-means, Hierarchical
- 15 Types of Regression you should know
LDA, QDA
Bagging
Chapter 8 of the book.
- Bootstrap mean is approximately a posterior average.
- Bootstrap aggregation or bagging average: Average the prediction over a collection of bootstrap samples, thereby reducing its variance. The bagging estimate is defined by
- [math]\displaystyle{ \hat{f}_{bag}(x) = \frac{1}{B}\sum_{b=1}^B \hat{f}^{*b}(x). }[/math]
Where Bagging Might Work Better Than Boosting
CLASSIFICATION FROM SCRATCH, BAGGING AND FORESTS 10/8
Boosting
- Ch8.2 Bagging, Random Forests and Boosting of An Introduction to Statistical Learning and the code.
- An Attempt To Understand Boosting Algorithm
- gbm package. An implementation of extensions to Freund and Schapire's AdaBoost algorithm and Friedman's gradient boosting machine. Includes regression methods for least squares, absolute loss, t-distribution loss, quantile regression, logistic, multinomial logistic, Poisson, Cox proportional hazards partial likelihood, AdaBoost exponential loss, Huberized hinge loss, and Learning to Rank measures (LambdaMart).
- https://www.biostat.wisc.edu/~kendzior/STAT877/illustration.pdf
- http://www.is.uni-freiburg.de/ressourcen/business-analytics/10_ensemblelearning.pdf and exercise
- Classification from scratch
AdaBoost
AdaBoost.M1 by Freund and Schapire (1997):
The error rate on the training sample is [math]\displaystyle{ \bar{err} = \frac{1}{N} \sum_{i=1}^N I(y_i \neq G(x_i)), }[/math]
Sequentially apply the weak classification algorithm to repeatedly modified versions of the data, thereby producing a sequence of weak classifiers [math]\displaystyle{ G_m(x), m=1,2,\dots,M. }[/math]
The predictions from all of them are combined through a weighted majority vote to produce the final prediction: [math]\displaystyle{ G(x) = sign[\sum_{m=1}^M \alpha_m G_m(x)]. }[/math] Here [math]\displaystyle{ \alpha_1,\alpha_2,\dots,\alpha_M }[/math] are computed by the boosting algorithm and weight the contribution of each respective [math]\displaystyle{ G_m(x) }[/math]. Their effect is to give higher influence to the more accurate classifiers in the sequence.
Dropout regularization
DART: Dropout Regularization in Boosting Ensembles
Gradient descent
Gradient descent is a first-order iterative optimization algorithm for finding the minimum of a function (Wikipedia).
- An Introduction to Gradient Descent and Linear Regression Easy to understand based on simple linear regression. Code is provided too.
- An overview of Gradient descent optimization algorithms
- A Complete Tutorial on Ridge and Lasso Regression in Python
- How to choose the learning rate?
- Machine learning from Andrew Ng
- http://scikit-learn.org/stable/modules/sgd.html
- R packages
The error function from a simple linear regression looks like
- [math]\displaystyle{ \begin{align} Err(m,b) &= \frac{1}{N}\sum_{i=1}^n (y_i - (m x_i + b))^2, \\ \end{align} }[/math]
We compute the gradient first for each parameters.
- [math]\displaystyle{ \begin{align} \frac{\partial Err}{\partial m} &= \frac{2}{n} \sum_{i=1}^n -x_i(y_i - (m x_i + b)), \\ \frac{\partial Err}{\partial b} &= \frac{2}{n} \sum_{i=1}^n -(y_i - (m x_i + b)) \end{align} }[/math]
The gradient descent algorithm uses an iterative method to update the estimates using a tuning parameter called learning rate.
new_m &= m_current - (learningRate * m_gradient) new_b &= b_current - (learningRate * b_gradient)
After each iteration, derivative is closer to zero. Coding in R for the simple linear regression.
Classification and Regression Trees (CART)
Construction of the tree classifier
- Node proportion
- [math]\displaystyle{ p(1|t) + \dots + p(6|t) =1 }[/math] where [math]\displaystyle{ p(j|t) }[/math] define the node proportions (class proportion of class j on node t. Here we assume there are 6 classes.
- Impurity of node t
- [math]\displaystyle{ i(t) }[/math] is a nonnegative function [math]\displaystyle{ \phi }[/math] of the [math]\displaystyle{ p(1|t), \dots, p(6|t) }[/math] such that [math]\displaystyle{ \phi(1/6,1/6,\dots,1/6) }[/math] = maximumm [math]\displaystyle{ \phi(1,0,\dots,0)=0, \phi(0,1,0,\dots,0)=0, \dots, \phi(0,0,0,0,0,1)=0 }[/math]. That is, the node impurity is largest when all classes are equally mixed together in it, and smallest when the node contains only one class.
- Gini index of impurity
- [math]\displaystyle{ i(t) = - \sum_{j=1}^6 p(j|t) \log p(j|t). }[/math]
- Goodness of the split s on node t
- [math]\displaystyle{ \Delta i(s, t) = i(t) -p_Li(t_L) - p_Ri(t_R). }[/math] where [math]\displaystyle{ p_R }[/math] are the proportion of the cases in t go into the left node [math]\displaystyle{ t_L }[/math] and a proportion [math]\displaystyle{ p_R }[/math] go into right node [math]\displaystyle{ t_R }[/math].
A tree was grown in the following way: At the root node [math]\displaystyle{ t_1 }[/math], a search was made through all candidate splits to find that split [math]\displaystyle{ s^* }[/math] which gave the largest decrease in impurity;
- [math]\displaystyle{ \Delta i(s^*, t_1) = \max_{s} \Delta i(s, t_1). }[/math]
- Class character of a terminal node was determined by the plurality rule. Specifically, if [math]\displaystyle{ p(j_0|t)=\max_j p(j|t) }[/math], then t was designated as a class [math]\displaystyle{ j_0 }[/math] terminal node.
R packages
Supervised Classification, Logistic and Multinomial
Variable selection and variable importance plot
Variable selection and cross-validation
- http://freakonometrics.hypotheses.org/19925
- http://ellisp.github.io/blog/2016/06/05/bootstrap-cv-strategies/
Mallow Cp
Mallows's Cp addresses the issue of overfitting. The Cp statistic calculated on a sample of data estimates the mean squared prediction error (MSPE).
- [math]\displaystyle{ E\sum_j (\hat{Y}_j - E(Y_j\mid X_j))^2/\sigma^2, }[/math]
The Cp statistic is defined as
- [math]\displaystyle{ C_p={SSE_p \over S^2} - N + 2P. }[/math]
- https://en.wikipedia.org/wiki/Mallows%27s_Cp
- Used in Yuan & Lin (2006) group lasso. The degrees of freedom is estimated by the bootstrap or perturbation methods. Their paper mentioned the performance is comparable with that of 5-fold CV but is computationally much faster.
Variable selection for mode regression
http://www.tandfonline.com/doi/full/10.1080/02664763.2017.1342781 Chen & Zhou, Journal of applied statistics ,June 2017
Neural network
- Build your own neural network in R
- (Video) 10.2: Neural Networks: Perceptron Part 1 - The Nature of Code from the Coding Train. The book THE NATURE OF CODE by DANIEL SHIFFMAN
- CLASSIFICATION FROM SCRATCH, NEURAL NETS. The ROCR package was used to produce the ROC curve.
Support vector machine (SVM)
- Improve SVM tuning through parallelism by using the foreach and doParallel packages.
Quadratic Discriminant Analysis (qda), KNN
Machine Learning. Stock Market Data, Part 3: Quadratic Discriminant Analysis and KNN
Regularization
Regularization is a process of introducing additional information in order to solve an ill-posed problem or to prevent overfitting
Ridge regression
- What is ridge regression?
- Why does ridge estimate become better than OLS by adding a constant to the diagonal? The estimates become more stable if the covariates are highly correlated.
- (In ridge regression) the matrix we need to invert no longer has determinant near zero, so the solution does not lead to uncomfortably large variance in the estimated parameters. And that’s a good thing. See this post.
Since L2 norm is used in the regularization, ridge regression is also called L2 regularization.
Hoerl and Kennard (1970a, 1970b) introduced ridge regression, which minimizes RSS subject to a constraint [math]\displaystyle{ \sum|\beta_j|^2 \le t }[/math]. Note that though ridge regression shrinks the OLS estimator toward 0 and yields a biased estimator [math]\displaystyle{ \hat{\beta} = (X^TX + \lambda X)^{-1} X^T y }[/math] where [math]\displaystyle{ \lambda=\lambda(t) }[/math], a function of t, the variance is smaller than that of the OLS estimator.
The solution exists if [math]\displaystyle{ \lambda \gt 0 }[/math] even if [math]\displaystyle{ n \lt p }[/math].
Ridge regression (L2 penalty) only shrinks the coefficients. In contrast, Lasso method (L1 penalty) tries to shrink some coefficient estimators to exactly zeros. This can be seen from comparing the coefficient path plot from both methods.
Geometrically (contour plot of the cost function), the L1 penalty (the sum of absolute values of coefficients) will incur a probability of some zero coefficients (i.e. some coefficient hitting the corner of a diamond shape in the 2D case). For example, in the 2D case (X-axis=[math]\displaystyle{ \beta_0 }[/math], Y-axis=[math]\displaystyle{ \beta_1 }[/math]), the shape of the L1 penalty [math]\displaystyle{ |\beta_0| + |\beta_1| }[/math] is a diamond shape whereas the shape of the L2 penalty ([math]\displaystyle{ \beta_0^2 + \beta_1^2 }[/math]) is a circle.
Lasso/glmnet and FAQs
- https://en.wikipedia.org/wiki/Lasso_(statistics). It has a discussion when two covariates are highly correlated. For example if gene [math]\displaystyle{ i }[/math] and gene [math]\displaystyle{ j }[/math] are identical, then the values of [math]\displaystyle{ \beta _{j} }[/math] and [math]\displaystyle{ \beta _{k} }[/math] that minimize the lasso objective function are not uniquely determined. Elastic Net has been designed to address this shortcoming.
- Strongly correlated covariates have similar regression coefficients, is referred to as the grouping effect. From the wikipedia page "one would like to find all the associated covariates, rather than selecting only one from each set of strongly correlated covariates, as lasso often does. In addition, selecting only a single covariate from each group will typically result in increased prediction error, since the model is less robust (which is why ridge regression often outperforms lasso)".
- Glmnet Vignette. It tries to minimize [math]\displaystyle{ RSS(\beta) + \lambda [(1-\alpha)||\beta||_2^2/2 + \alpha ||\beta||_1] }[/math]. The elastic-net penalty is controlled by [math]\displaystyle{ \alpha }[/math], and bridge the gap between lasso ([math]\displaystyle{ \alpha = 1 }[/math]) and ridge ([math]\displaystyle{ \alpha = 0 }[/math]). Following is a CV curve (adaptive lasso) using the example from glmnet(). Two vertical lines are indicated: left one is lambda.min (that gives minimum mean cross-validated error) and right one is lambda.1se (the most regularized model such that error is within one standard error of the minimum). For the tuning parameter [math]\displaystyle{ \lambda }[/math],
- The larger the [math]\displaystyle{ \lambda }[/math], more coefficients are becoming zeros (think about coefficient path plots) and thus the simpler (more regularized) the model.
- If [math]\displaystyle{ \lambda }[/math] becomes zero, it reduces to the regular regression and if [math]\displaystyle{ \lambda }[/math] becomes infinity, the coefficients become zeros.
- In terms of the bias-variance tradeoff, the larger the [math]\displaystyle{ \lambda }[/math], the higher the bias and the lower the variance of the coefficient estimators.
set.seed(1010)
n=1000;p=100
nzc=trunc(p/10)
x=matrix(rnorm(n*p),n,p)
beta=rnorm(nzc)
fx= x[,seq(nzc)] %*% beta
eps=rnorm(n)*5
y=drop(fx+eps)
px=exp(fx)
px=px/(1+px)
ly=rbinom(n=length(px),prob=px,size=1)
## Full lasso
set.seed(999)
cv.full <- cv.glmnet(x, ly, family='binomial', alpha=1, parallel=TRUE)
plot(cv.full) # cross-validation curve and two lambda's
plot(glmnet(x, ly, family='binomial', alpha=1), xvar="lambda", label=TRUE) # coefficient path plot
plot(glmnet(x, ly, family='binomial', alpha=1)) # L1 norm plot
log(cv.full$lambda.min) # -4.546394
log(cv.full$lambda.1se) # -3.61605
sum(coef(cv.full, s=cv.full$lambda.min) != 0) # 44
## Ridge Regression to create the Adaptive Weights Vector
set.seed(999)
cv.ridge <- cv.glmnet(x, ly, family='binomial', alpha=0, parallel=TRUE)
wt <- 1/abs(matrix(coef(cv.ridge, s=cv.ridge$lambda.min)
[, 1][2:(ncol(x)+1)] ))^1 ## Using gamma = 1, exclude intercept
## Adaptive Lasso using the 'penalty.factor' argument
set.seed(999)
cv.lasso <- cv.glmnet(x, ly, family='binomial', alpha=1, parallel=TRUE, penalty.factor=wt)
# defautl type.measure="deviance" for logistic regression
plot(cv.lasso)
log(cv.lasso$lambda.min) # -2.995375
log(cv.lasso$lambda.1se) # -0.7625655
sum(coef(cv.lasso, s=cv.lasso$lambda.min) != 0) # 34
- A list of potential lambdas: see Linear Regression case. The λ sequence is determined by lambda.max and lambda.min.ratio. The latter (default is ifelse(nobs<nvars,0.01,0.0001)) is the ratio of smallest value of the generated λ sequence (say lambda.min) to lambda.max. The program then generated nlambda values linear on the log scale from lambda.max down to lambda.min. lambda.max is not given, but easily computed from the input x and y; it is the smallest value for lambda such that all the coefficients are zero.
- lambda.min vs lambda.1se
- The lambda.1se represents the value of λ in the search that was simpler than the best model (lambda.min), but which has error within 1 standard error of the best model. In other words, using the value of lambda.1se as the selected value for λ results in a model that is slightly simpler than the best model but which cannot be distinguished from the best model in terms of error given the uncertainty in the k-fold CV estimate of the error of the best model.
- The lambda.min option refers to value of λ at the lowest CV error. The error at this value of λ is the average of the errors over the k folds and hence this estimate of the error is uncertain.
- https://www.rdocumentation.org/packages/glmnet/versions/2.0-10/topics/glmnet
- glmnetUtils: quality of life enhancements for elastic net regression with glmnet
- When the LASSO fails???
- Mixing parameter: alpha=1 is the lasso penalty, and alpha=0 the ridge penalty and anything between 0–1 is Elastic net.
- RIdge regression uses Euclidean distance/L2-norm as the penalty. It won't remove any variables.
- Lasso uses L1-norm as the penalty. Some of the coefficients may be shrunk exactly to zero.
- In ridge regression and lasso, what is lambda?
- Lambda is a penalty coefficient. Large lambda will shrink the coefficients.
- cv.glment()$lambda.1se gives the most regularized model such that error is within one standard error of the minimum
- cv.glmnet() has a logical parameter parallel which is useful if a cluster or cores have been previously allocated
- Ridge regression and the LASSO
- Standard error/Confidence interval
- Standard Errors in GLMNET and Confidence intervals for Ridge regression
- Why SEs are not meaningful and are usually not provided in penalized regression?
- Hint: standard errors are not very meaningful for strongly biased estimates such as arise from penalized estimation methods.
- Penalized estimation is a procedure that reduces the variance of estimators by introducing substantial bias.
- The bias of each estimator is therefore a major component of its mean squared error, whereas its variance may contribute only a small part.
- Any bootstrap-based calculations can only give an assessment of the variance of the estimates.
- Reliable estimates of the bias are only available if reliable unbiased estimates are available, which is typically not the case in situations in which penalized estimates are used.
- Hottest glmnet questions from stackexchange.
- Standard errors for lasso prediction. There might not be a consensus on a statistically valid method of calculating standard errors for the lasso predictions.
- Code for Adaptive-Lasso for Cox's proportional hazards model by Zhang & Lu (2007). This can compute the SE of estimates. The weights are originally based on the maximizers of the log partial likelihood. However, the beta may not be estimable in cases such as high-dimensional gene data, or the beta may be unstable if strong collinearity exists among covariates. In such cases, robust estimators such as ridge regression estimators would be used to determine the weights.
- LASSO vs Least angle regression
- https://web.stanford.edu/~hastie/Papers/LARS/LeastAngle_2002.pdf
- Least Angle Regression, Forward Stagewise and the Lasso
- https://www.quora.com/What-is-Least-Angle-Regression-and-when-should-it-be-used
- A simple explanation of the Lasso and Least Angle Regression
- https://stats.stackexchange.com/questions/4663/least-angle-regression-vs-lasso
- https://cran.r-project.org/web/packages/lars/index.html
- Oracle property and adaptive lasso
- Variable Selection via Nonconcave Penalized Likelihood and Its Oracle Properties, Fan & Li (2001) JASA
- Adaptive Lasso: What it is and how to implement in R. Adaptive lasso weeks to minimize [math]\displaystyle{ RSS(\beta) + \lambda \sum_1^p \hat{\omega}_j |\beta_j| }[/math] where [math]\displaystyle{ \lambda }[/math] is the tuning parameter, [math]\displaystyle{ \hat{\omega}_j = \frac{1}{(|\hat{\beta}_j^{ini}|)^\gamma} }[/math] is the adaptive weights vector and [math]\displaystyle{ \hat{\beta}_j^{ini} }[/math] is an initial estimate of the coefficients obtained through ridge regression. Adaptive Lasso ends up penalizing more those coefficients with lower initial estimates. [math]\displaystyle{ \gamma }[/math] is a positive constant for adjustment of the adaptive weight vector, and the authors suggest the possible values of 0.5, 1 and 2.
- When n goes to infinity, [math]\displaystyle{ \hat{\omega}_j |\beta_j| }[/math] converges to [math]\displaystyle{ I(\beta_j \neq 0) }[/math]. So the adaptive Lasso procedure can be regarded as an automatic implementation of best-subset selection in some asymptotic sense.
- What is the oracle property of an estimator? An oracle estimator must be consistent in 1) variable selection and 2) consistent parameter estimation.
- (Linear regression) The adaptive lasso and its oracle properties Zou (2006, JASA)
- (Cox model) Adaptive-LASSO for Cox's proportional hazard model by Zhang and Lu (2007, Biometrika)
- Some issues:
- With group of highly correlated features, Lasso tends to select amongst them arbitrarily.
- Often empirically ridge has better predictive performance than lasso but lasso leads to sparser solution
- Elastic-net (Zou & Hastie '05) aims to address these issues: hybrid between Lasso and ridge regression, uses L1 and L2 penalties.
Lasso logistic regression
https://freakonometrics.hypotheses.org/52894
How to solve lasso/convex optimization
- Convex Optimization by Boyd S, Vandenberghe L, Cambridge 2004. It is cited by Zhang & Lu (2007). The interior point algorithm can be used to solve the optimization problem in adaptive lasso.
- Review of gradient descent:
- Finding maximum: [math]\displaystyle{ w^{(t+1)} = w^{(t)} + \eta \frac{dg(w)}{dw} }[/math], where [math]\displaystyle{ \eta }[/math] is stepsize.
- Finding minimum: [math]\displaystyle{ w^{(t+1)} = w^{(t)} - \eta \frac{dg(w)}{dw} }[/math].
- What is the difference between Gradient Descent and Newton's Gradient Descent? Newton's method requires [math]\displaystyle{ g''(w) }[/math], more smoothness of g(.).
- Finding minimum for multiple variables (gradient descent): [math]\displaystyle{ w^{(t+1)} = w^{(t)} - \eta \Delta g(w^{(t)}) }[/math]. For the least squares problem, [math]\displaystyle{ g(w) = RSS(w) }[/math].
- Finding minimum for multiple variables in the least squares problem (minimize [math]\displaystyle{ RSS(w) }[/math]): [math]\displaystyle{ \text{partial}(j) = -2\sum h_j(x_i)(y_i - \hat{y}_i(w^{(t)}), w_j^{(t+1)} = w_j^{(t)} - \eta \; \text{partial}(j) }[/math]
- Finding minimum for multiple variables in the ridge regression problem (minimize [math]\displaystyle{ RSS(w)+\lambda ||w||_2^2=(y-Hw)'(y-Hw)+\lambda w'w }[/math]): [math]\displaystyle{ \text{partial}(j) = -2\sum h_j(x_i)(y_i - \hat{y}_i(w^{(t)}), w_j^{(t+1)} = (1-2\eta \lambda) w_j^{(t)} - \eta \; \text{partial}(j) }[/math]. Compared to the closed form approach: [math]\displaystyle{ \hat{w} = (H'H + \lambda I)^{-1}H'y }[/math] where 1. the inverse exists even N<D as long as [math]\displaystyle{ \lambda \gt 0 }[/math] and 2. the complexity of inverse is [math]\displaystyle{ O(D^3) }[/math], D is the dimension of the covariates.
- Cyclical coordinate descent was used (vignette) in the glmnet package. See also coordinate descent. The reason we call it 'descent' is because we want to 'minimize' an objective function.
- [math]\displaystyle{ \hat{w}_j = \min_w g(\hat{w}_1, \cdots, \hat{w}_{j-1},w, \hat{w}_{j+1}, \cdots, \hat{w}_D) }[/math]
- See paper on JSS 2010. The Cox PHM case also uses the cyclical coordinate descent method; see the paper on JSS 2011.
- Coursera's Machine learning course 2: Regression at 1:42. Soft-thresholding the coefficients is the key for the L1 penalty. The range for the thresholding is controlled by [math]\displaystyle{ \lambda }[/math]. Note to view the videos and all materials in coursera we can enroll to audit the course without starting a trial.
- No step size is required as in gradient descent.
- Implementing LASSO Regression with Coordinate Descent, Sub-Gradient of the L1 Penalty and Soft Thresholding in Python
- Coordinate descent in the least squares problem: [math]\displaystyle{ \frac{\partial}{\partial w_j} RSS(w)= -2 \rho_j + 2 w_j }[/math]; i.e. [math]\displaystyle{ \hat{w}_j = \rho_j }[/math].
- Coordinate descent in the Lasso problem (for normalized features): [math]\displaystyle{ \hat{w}_j = \begin{cases} \rho_j + \lambda/2, & \text{if }\rho_j \lt -\lambda/2 \\ 0, & \text{if } -\lambda/2 \le \rho_j \le \lambda/2\\ \rho_j- \lambda/2, & \text{if }\rho_j \gt \lambda/2 \end{cases} }[/math]
- Choosing [math]\displaystyle{ \lambda }[/math] via cross validation tends to favor less sparse solutions and thus smaller [math]\displaystyle{ \lambda }[/math] then optimal choice for feature selection. See "Machine learning: a probabilistic perspective", Murphy 2012.
- Classical: Least angle regression (LARS) Efron et al 2004.
- Alternating Direction Method of Multipliers (ADMM). Boyd, 2011. “Distributed Optimization and Statistical Learning via the Alternating Direction Method of Multipliers.” Foundations and Trends in Machine Learning. Vol. 3, No. 1, 2010, pp. 1–122.
- If some variables in design matrix are correlated, then LASSO is convex or not?
- Tibshirani. Regression shrinkage and selection via the lasso (free). JRSS B 1996.
- Convex Optimization in R by Koenker & Mizera 2014.
- Pathwise coordinate optimization by Friedman et al 2007.
- Statistical learning with sparsity: the Lasso and generalizations T. Hastie, R. Tibshirani, and M. Wainwright, 2015 (book)
- Element of Statistical Learning (book)
- https://youtu.be/A5I1G1MfUmA StatsLearning Lect8h 110913
- Fu's (1998) shooting algorithm for Lasso (mentioned in the history of coordinate descent) and Zhang & Lu's (2007) modified shooting algorithm for adaptive Lasso.
- Machine Learning: a Probabilistic Perspective Choosing [math]\displaystyle{ \lambda }[/math] via cross validation tends to favor less sparse solutions and thus smaller [math]\displaystyle{ \lambda }[/math] than optimal choice for feature selection.
Quadratic programming
- https://en.wikipedia.org/wiki/Quadratic_programming
- https://en.wikipedia.org/wiki/Lasso_(statistics)
- CRAN Task View: Optimization and Mathematical Programming
- quadprog package and solve.QP() function
- Solving Quadratic Progams with R’s quadprog package
- More on Quadratic Programming in R
- https://optimization.mccormick.northwestern.edu/index.php/Quadratic_programming
- Maximin projection learning for optimal treatment decision with heterogeneous individualized treatment effects where the algorithm from Lee 2016 was used.
1. Elastic net
2. Group lasso
- Yuan and Lin 2006 JRSSB
- https://cran.r-project.org/web/packages/gglasso/, http://royr2.github.io/2014/04/15/GroupLasso.html
- https://cran.r-project.org/web/packages/grpreg/
- https://cran.r-project.org/web/packages/grplasso/ by Lukas Meier (paper), used in the biospear package for survival data
- https://cran.r-project.org/web/packages/SGL/index.html, http://royr2.github.io/2014/05/20/SparseGroupLasso.html, http://web.stanford.edu/~hastie/Papers/SGLpaper.pdf
Comparison by plotting
If we are running simulation, we can use the DALEX package to visualize the fitting result from different machine learning methods and the true model. See http://smarterpoland.pl/index.php/2018/05/ml-models-what-they-cant-learn.
Imbalanced Classification
Deep Learning
- CS294-129 Designing, Visualizing and Understanding Deep Neural Networks from berkeley.
- https://www.youtube.com/playlist?list=PLkFD6_40KJIxopmdJF_CLNqG3QuDFHQUm
- Deep Learning from first principles in Python, R and Octave – Part 5
Tensor Flow (tensorflow package)
- https://tensorflow.rstudio.com/
- Machine Learning with R and TensorFlow (Video)
- Machine Learning Crash Course with TensorFlow APIs
- Predicting cancer outcomes from histology and genomics using convolutional networks Pooya Mobadersany et al, PNAS 2018
Biological applications
Machine learning resources
These Machine Learning Courses Will Prepare a Career Path for You
Randomization inference
- Google: randomization inference in r
- Randomization Inference for Outcomes with Clumping at Zero, The American Statistician 2018
- Randomization inference vs. bootstrapping for p-values
Bootstrap
- Bootstrapping made easy and tidy with slipper
- bootstrap package. An Introduction to the Bootstrap" by B. Efron and R. Tibshirani, 1993
- boot package. Functions and datasets for bootstrapping from the book "Bootstrap Methods and Their Application" by A. C. Davison and D. V. Hinkley (1997, CUP). The main functions are boot() and boot.ci().
- https://www.rdocumentation.org/packages/boot/versions/1.3-20
- https://www.statmethods.net/advstats/bootstrapping.html Nonparametric bootstrapping
- http://people.tamu.edu/~alawing/materials/ESSM689/Btutorial.pdf
- http://statweb.stanford.edu/~tibs/sta305files/FoxOnBootingRegInR.pdf
- http://www.stat.wisc.edu/~larget/stat302/chap3.pdf
- http://www.math.ntu.edu.tw/~hchen/teaching/LargeSample/references/R-bootstrap.pdf & http://www.math.ntu.edu.tw/~hchen/teaching/LargeSample/notes/notebootstrap.pdf No package is used
- http://web.as.uky.edu/statistics/users/pbreheny/621/F10/notes/9-21.pdf Bootstrap confidence interval
- http://www-stat.wharton.upenn.edu/~stine/research/spida_2005.pdf
Cross Validation
R packages:
- rsample (released July 2017)
- CrossValidate (released July 2017)
.632 bootstrap
Create partitions
set.seed(), sample.split(),createDataPartition(), and createFolds() functions.
k-fold cross validation with modelr and broom
Nested resampling
- Nested Resampling with rsample
- https://stats.stackexchange.com/questions/292179/whats-the-meaning-of-nested-resampling
Nested resampling is need when we want to tuning a model by using a grid search. The default settings of a model are likely not optimal for each data set out. So an inner CV has to be performed with the aim to find the best parameter set of a learner for each fold.
See a diagram at https://i.stack.imgur.com/vh1sZ.png
In BRB-ArrayTools -> class prediction with multiple methods, the alpha (significant level of threshold used for gene selection, 2nd option in individual genes) can be viewed as a tuning parameter for the development of a classifier.
Pre-validation
- Pre-validation and inference in microarrays Statistical Applications in Genetics and Molecular Biology, 2002.
- http://www.stat.columbia.edu/~tzheng/teaching/genetics/papers/tib_efron.pdf
Clustering
k-means clustering
- Assumptions, a post from varianceexplained.org.
Hierarchical clustering
For the kth cluster, define the Error Sum of Squares as [math]\displaystyle{ ESS_m = }[/math] sum of squared deviations (squared Euclidean distance) from the cluster centroid. [math]\displaystyle{ ESS_m = \sum_{l=1}^{n_m}\sum_{k=1}^p (x_{ml,k} - \bar{x}_{m,k})^2 }[/math] in which [math]\displaystyle{ \bar{x}_{m,k} = (1/n_m) \sum_{l=1}^{n_m} x_{ml,k} }[/math] the mean of the mth cluster for the kth variable, [math]\displaystyle{ x_{ml,k} }[/math] being the score on the kth variable [math]\displaystyle{ (k=1,\dots,p) }[/math] for the lth object [math]\displaystyle{ (l=1,\dots,n_m) }[/math] in the mth cluster [math]\displaystyle{ (m=1,\dots,g) }[/math].
If there are C clusters, define the Total Error Sum of Squares as Sum of Squares as [math]\displaystyle{ ESS = \sum_m ESS_m, m=1,\dots,C }[/math]
Consider the union of every possible pair of clusters.
Combine the 2 clusters whose combination combination results in the smallest increase in ESS.
Comments:
- Ward's method tends to join clusters with a small number of observations, and it is strongly biased toward producing clusters with the same shape and with roughly the same number of observations.
- It is also very sensitive to outliers. See Milligan (1980).
Take pomeroy data (7129 x 90) for an example:
library(gplots)
lr = read.table("C:/ArrayTools/Sample datasets/Pomeroy/Pomeroy -Project/NORMALIZEDLOGINTENSITY.txt")
lr = as.matrix(lr)
method = "average" # method <- "complete"; method <- "ward.D"; method <- "ward.D2"
hclust1 <- function(x) hclust(x, method= method)
heatmap.2(lr, col=bluered(75), hclustfun = hclust1, distfun = dist,
density.info="density", scale = "none",
key=FALSE, symkey=FALSE, trace="none",
main = method)
Density based clustering
http://www.r-exercises.com/2017/06/10/density-based-clustering-exercises/
Optimal number of clusters
Mixed Effect Model
- Paper by Laird and Ware 1982
- John Fox's Linear Mixed Models Appendix to An R and S-PLUS Companion to Applied Regression. Very clear. It provides 2 typical examples (hierarchical data and longitudinal data) of using the mixed effects model. It also uses Trellis plots to examine the data.
- Chapter 10 Random and Mixed Effects from Modern Applied Statistics with S by Venables and Ripley.
- (Book) lme4: Mixed-effects modeling with R by Douglas Bates.
- (Book) Mixed-effects modeling in S and S-Plus by José Pinheiro and Douglas Bates.
- Simulation and power analysis of generalized linear mixed models
- Linear mixed-effect models in R by poissonisfish
Model selection criteria
- Assessing the Accuracy of our models (R Squared, Adjusted R Squared, RMSE, MAE, AIC)
- Comparing additive and multiplicative regressions using AIC in R
Overfitting
How to judge if a supervised machine learning model is overfitting or not?
Entropy
Definition
Entropy is defined by -log2(p) where p is a probability. Higher entropy represents higher unpredictable of an event.
Some examples:
- Fair 2-side die: Entropy = -.5*log2(.5) - .5*log2(.5) = 1.
- Fair 6-side die: Entropy = -6*1/6*log2(1/6) = 2.58
- Weighted 6-side die: Consider pi=.1 for i=1,..,5 and p6=.5. Entropy = -5*.1*log2(.1) - .5*log2(.5) = 2.16 (less unpredictable than a fair 6-side die).
Use
When entropy was applied to the variable selection, we want to select a class variable which gives a largest entropy difference between without any class variable (compute entropy using response only) and with that class variable (entropy is computed by adding entropy in each class level) because this variable is most discriminative and it gives most information gain. For example,
- entropy (without any class)=.94,
- entropy(var 1) = .69,
- entropy(var 2)=.91,
- entropy(var 3)=.725.
We will choose variable 1 since it gives the largest gain (.94 - .69) compared to the other variables (.94 -.91, .94 -.725).
Why is picking the attribute with the most information gain beneficial? It reduces entropy, which increases predictability. A decrease in entropy signifies an decrease in unpredictability, which also means an increase in predictability.
Consider a split of a continuous variable. Where should we cut the continuous variable to create a binary partition with the highest gain? Suppose cut point c1 creates an entropy .9 and another cut point c2 creates an entropy .1. We should choose c2.
Related
In addition to information gain, gini (dʒiːni) index is another metric used in decision tree. See wikipedia page about decision tree learning.
Ensembles
Combining classifiers. Pro: better classification performance. Con: time consuming.
Comic http://flowingdata.com/2017/09/05/xkcd-ensemble-model/
Bagging
Draw N bootstrap samples and summary the results (averaging for regression problem, majority vote for classification problem). Decrease variance without changing bias. Not help much with underfit or high bias models.
Random forest
Variance importance: if you scramble the values of a variable, and the accuracy of your tree does not change much, then the variable is not very important.
Why is it useful to compute variance importance? So the model's predictions are easier to interpret (not improve the prediction performance).
Random forest has advantages of easier to run in parallel and suitable for small n large p problems.
Random forest versus logistic regression: a large-scale benchmark experiment by Raphael Couronné, BMC Bioinformatics 2018
Boosting
Instead of selecting data points randomly with the boostrap, it favors the misclassified points.
Algorithm:
- Initialize the weights
- Repeat
- resample with respect to weights
- retrain the model
- recompute weights
Since boosting requires computation in iterative and bagging can be run in parallel, bagging has an advantage over boosting when the data is very large.
Time series
Ensemble learning for time series forecasting in R
p-values
p-values
- Prob(Data | H0)
- https://en.wikipedia.org/wiki/P-value
- THE ASA SAYS NO TO P-VALUES The problem is that with large samples, significance tests pounce on tiny, unimportant departures from the null hypothesis. We have the opposite problem with small samples: The power of the test is low, and we will announce that there is “no significant effect” when in fact we may have too little data to know whether the effect is important.
- It’s not the p-values’ fault
- Exploring P-values with Simulations in R from Stable Markets.
- p-value and effect size. http://journals.sagepub.com/doi/full/10.1177/1745691614553988
Distribution of p values in medical abstracts
- http://www.ncbi.nlm.nih.gov/pubmed/26608725
- An R package with several million published p-values in tidy data sets by Jeff Leek.
nominal p-value and Empirical p-values
- Nominal p-values are based on asymptotic null distributions
- Empirical p-values are computed from simulations/permutations
(nominal) alpha level
Conventional methodology for statistical testing is, in advance of undertaking the test, to set a NOMINAL ALPHA CRITERION LEVEL (often 0.05). The outcome is classified as showing STATISTICAL SIGNIFICANCE if the actual ALPHA (probability of the outcome under the null hypothesis) is no greater than this NOMINAL ALPHA CRITERION LEVEL.
- http://www.translationdirectory.com/glossaries/glossary033.htm
- http://courses.washington.edu/p209s07/lecturenotes/Week%205_Monday%20overheads.pdf
T-statistic
Let [math]\displaystyle{ \scriptstyle\hat\beta }[/math] be an estimator of parameter β in some statistical model. Then a t-statistic for this parameter is any quantity of the form
- [math]\displaystyle{ t_{\hat{\beta}} = \frac{\hat\beta - \beta_0}{\mathrm{s.e.}(\hat\beta)}, }[/math]
where β0 is a non-random, known constant, and [math]\displaystyle{ \scriptstyle s.e.(\hat\beta) }[/math] is the standard error of the estimator [math]\displaystyle{ \scriptstyle\hat\beta }[/math].
Two sample test assuming equal variance
The t statistic to test whether the means are different can be calculated as follows:
- [math]\displaystyle{ t = \frac{\bar {X}_1 - \bar{X}_2}{s_{X_1X_2} \cdot \sqrt{\frac{1}{n_1}+\frac{1}{n_2}}} }[/math]
where
- [math]\displaystyle{ s_{X_1X_2} = \sqrt{\frac{(n_1-1)s_{X_1}^2+(n_2-1)s_{X_2}^2}{n_1+n_2-2}}. }[/math]
[math]\displaystyle{ s_{X_1X_2} }[/math] is an estimator of the common/pooled standard deviation of the two samples. The square-root of a pooled variance estimator is known as a pooled standard deviation.
The degrees of freedom is :[math]\displaystyle{ n_1 + n_2 - 2. }[/math]
Two sample test assuming unequal variance
The t statistic (Behrens-Welch test statistic) to test whether the population means are different is calculated as:
- [math]\displaystyle{ t = {\overline{X}_1 - \overline{X}_2 \over s_{\overline{X}_1 - \overline{X}_2}} }[/math]
where
- [math]\displaystyle{ s_{\overline{X}_1 - \overline{X}_2} = \sqrt{{s_1^2 \over n_1} + {s_2^2 \over n_2}}. }[/math]
Here s2 is the unbiased estimator of the variance of the two samples.
The degrees of freedom is evaluated using the Satterthwaite's approximation
- [math]\displaystyle{ df = { ({s_1^2 \over n_1} + {s_2^2 \over n_2})^2 \over {({s_1^2 \over n_1})^2 \over n_1-1} + {({s_2^2 \over n_2})^2 \over n_2-1} }. }[/math]
Unpooled vs pooled methods
Paired test
Have you ever asked yourself, "how should I approach the classic pre-post analysis?"
Z-value/Z-score
If the population parameters are known, then rather than computing the t-statistic, one can compute the z-score.
Nonparametric test: Wilcoxon rank sum test
Sensitive to differences in location
Nonparametric test: Kolmogorov-Smirnov test
Sensitive to difference in shape and location of the distribution functions of two groups
Limma: Empirical Bayes method
- Some Bioconductor packages: limma, RnBeads, IMA, minfi packages.
- The moderated T-statistics used in Limma is defined on Limma's user guide.
- Correct assumptions of using limma moderated t-test and the paper Should We Abandon the t-Test in the Analysis of Gene Expression Microarray Data: A Comparison of Variance Modeling Strategies.
- Evaluation: statistical power (figure 3, 4, 5), false-positive rate (table 2), execution time and ease of use (table 3)
- Limma presents several advantages
- RVM inflates the expected number of false-positives when sample size is small. On the other hand the, RVM is very close to Limma from either their formulas (p3 of the supporting info) or the Hierarchical clustering (figure 2) of two examples.
ANOVA
- A simple ANOVA
- Repeated measures ANOVA in R Exercises
- Mixed models for ANOVA designs with one observation per unit of observation and cell of the design
- afex package
- Experiment designs for Agriculture
TukeyHSD, diagnostic checking
https://datascienceplus.com/one-way-anova-in-r/
- TukeyHSD for the pairwise tests
- Shapiro-Wilk test for normality
- Bartlett test and Levene test for the homogeneity of variances across the groups
Repeated measure
Sample Size
- Why Within-Subject Designs Require Fewer Participants than Between-Subject Designs
- Calculating required sample size in R and SAS
- Power analysis and sample size calculation for Agriculture
Goodness of fit
Chi-square tests
Fitting distribution
Contingency Tables
Odds ratio and Risk ratio
The ratio of the odds of an event occurring in one group to the odds of it occurring in another group
drawn | not drawn | ------------------------------------- white | A | B | Wh ------------------------------------- black | C | D | Bk
- Odds Ratio = (A / C) / (B / D) = (AD) / (BC)
- Risk Ratio = (A / Wh) / (C / Bk)
Hypergeometric, One-tailed Fisher exact test
- https://www.bioconductor.org/help/course-materials/2009/SeattleApr09/gsea/ (Are interesting features over-represented? or are selected genes more often in the GO category than expected by chance?)
- https://en.wikipedia.org/wiki/Hypergeometric_distribution. In a test for over-representation of successes in the sample, the hypergeometric p-value is calculated as the probability of randomly drawing k or more successes from the population in n total draws. In a test for under-representation, the p-value is the probability of randomly drawing k or fewer successes.
- http://stats.stackexchange.com/questions/62235/one-tailed-fishers-exact-test-and-the-hypergeometric-distribution
- Two sided hypergeometric test
- https://www.biostars.org/p/90662/ When computing the p-value (tail probability), consider to use 1 - Prob(observed -1) instead of 1 - Prob(observed) for discrete distribution.
- https://stat.ethz.ch/R-manual/R-devel/library/stats/html/Hypergeometric.html p(x) = choose(m, x) choose(n, k-x) / choose(m+n, k).
drawn | not drawn |
-------------------------------------
white | x | | m
-------------------------------------
black | k-x | | n
-------------------------------------
| k | | m+n
For example, k=100, m=100, m+n=1000,
> 1 - phyper(10, 100, 10^3-100, 100, log.p=F) [1] 0.4160339 > a <- dhyper(0:10, 100, 10^3-100, 100) > cumsum(rev(a)) [1] 1.566158e-140 1.409558e-135 3.136408e-131 3.067025e-127 1.668004e-123 5.739613e-120 1.355765e-116 [8] 2.325536e-113 3.018276e-110 3.058586e-107 2.480543e-104 1.642534e-101 9.027724e-99 4.175767e-96 [15] 1.644702e-93 5.572070e-91 1.638079e-88 4.210963e-86 9.530281e-84 1.910424e-81 3.410345e-79 [22] 5.447786e-77 7.821658e-75 1.013356e-72 1.189000e-70 1.267638e-68 1.231736e-66 1.093852e-64 [29] 8.900857e-63 6.652193e-61 4.576232e-59 2.903632e-57 1.702481e-55 9.240350e-54 4.650130e-52 [36] 2.173043e-50 9.442985e-49 3.820823e-47 1.441257e-45 5.074077e-44 1.669028e-42 5.134399e-41 [43] 1.478542e-39 3.989016e-38 1.009089e-36 2.395206e-35 5.338260e-34 1.117816e-32 2.200410e-31 [50] 4.074043e-30 7.098105e-29 1.164233e-27 1.798390e-26 2.617103e-25 3.589044e-24 4.639451e-23 [57] 5.654244e-22 6.497925e-21 7.042397e-20 7.198582e-19 6.940175e-18 6.310859e-17 5.412268e-16 [64] 4.377256e-15 3.338067e-14 2.399811e-13 1.626091e-12 1.038184e-11 6.243346e-11 3.535115e-10 [71] 1.883810e-09 9.442711e-09 4.449741e-08 1.970041e-07 8.188671e-07 3.193112e-06 1.167109e-05 [78] 3.994913e-05 1.279299e-04 3.828641e-04 1.069633e-03 2.786293e-03 6.759071e-03 1.525017e-02 [85] 3.196401e-02 6.216690e-02 1.120899e-01 1.872547e-01 2.898395e-01 4.160339e-01 5.550192e-01 [92] 6.909666e-01 8.079129e-01 8.953150e-01 9.511926e-01 9.811343e-01 9.942110e-01 9.986807e-01 [99] 9.998018e-01 9.999853e-01 1.000000e+00 # Density plot plot(0:100, dhyper(0:100, 100, 10^3-100, 100), type='h')
Moreover,
1 - phyper(q=10, m, n, k)
= 1 - sum_{x=0}^{x=10} phyper(x, m, n, k)
= 1 - sum(a[1:11]) # R's index starts from 1.
Another example is the data from the functional annotation tool in DAVID.
| gene list | not gene list |
-------------------------------------------------------
pathway | 3 (q) | | 40 (m)
-------------------------------------------------------
not in pathway | 297 | | 29960 (n)
-------------------------------------------------------
| 300 (k) | | 30000
The one-tailed p-value from the hypergeometric test is calculated as 1 - phyper(3-1, 40, 29960, 300) = 0.0074.
Fisher's exact test
Following the above example from the DAVID website, the following R command calculates the Fisher exact test for independence in 2x2 contingency tables.
> fisher.test(matrix(c(3, 40, 297, 29960), nr=2)) # alternative = "two.sided" by default
Fisher's Exact Test for Count Data
data: matrix(c(3, 40, 297, 29960), nr = 2)
p-value = 0.008853
alternative hypothesis: true odds ratio is not equal to 1
95 percent confidence interval:
1.488738 23.966741
sample estimates:
odds ratio
7.564602
> fisher.test(matrix(c(3, 40, 297, 29960), nr=2), alternative="greater")
Fisher's Exact Test for Count Data
data: matrix(c(3, 40, 297, 29960), nr = 2)
p-value = 0.008853
alternative hypothesis: true odds ratio is greater than 1
95 percent confidence interval:
1.973 Inf
sample estimates:
odds ratio
7.564602
> fisher.test(matrix(c(3, 40, 297, 29960), nr=2), alternative="less")
Fisher's Exact Test for Count Data
data: matrix(c(3, 40, 297, 29960), nr = 2)
p-value = 0.9991
alternative hypothesis: true odds ratio is less than 1
95 percent confidence interval:
0.00000 20.90259
sample estimates:
odds ratio
7.564602
From the documentation of fisher.test
Usage:
fisher.test(x, y = NULL, workspace = 200000, hybrid = FALSE,
control = list(), or = 1, alternative = "two.sided",
conf.int = TRUE, conf.level = 0.95,
simulate.p.value = FALSE, B = 2000)
- For 2 by 2 cases, p-values are obtained directly using the (central or non-central) hypergeometric distribution.
- For 2 by 2 tables, the null of conditional independence is equivalent to the hypothesis that the odds ratio equals one.
- The alternative for a one-sided test is based on the odds ratio, so ‘alternative = "greater"’ is a test of the odds ratio being bigger than ‘or’.
- Two-sided tests are based on the probabilities of the tables, and take as ‘more extreme’ all tables with probabilities less than or equal to that of the observed table, the p-value being the sum of such probabilities.
Chi-square independence test
Exploring the underlying theory of the chi-square test through simulation - part 2
GSEA
Determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states
- https://www.bioconductor.org/help/course-materials/2015/SeattleApr2015/E_GeneSetEnrichment.html
- http://software.broadinstitute.org/gsea/index.jsp
- Statistical power of gene-set enrichment analysis is a function of gene set correlation structure by SWANSON 2017
Two categories of GSEA procedures:
- Competitive: compare genes in the test set relative to all other genes.
- Self-contained: whether the gene-set is more DE than one were to expect under the null of no association between two phenotype conditions (without reference to other genes in the genome). For example the method by Jiang & Gentleman Bioinformatics 2007
Confidence vs Credibility Intervals
http://freakonometrics.hypotheses.org/18117
Power Analysis
Power analysis for default Bayesian t-tests
http://daniellakens.blogspot.com/2016/01/power-analysis-for-default-bayesian-t.html
Using simulation for power analysis: an example based on a stepped wedge study design
https://www.rdatagen.net/post/using-simulation-for-power-analysis-an-example/
Power analysis and sample size calculation for Agriculture
http://r-video-tutorial.blogspot.com/2017/07/power-analysis-and-sample-size.html
Power calculation for proportions (shiny app)
https://juliasilge.shinyapps.io/power-app/
Common covariance/correlation structures
See psu.edu. Assume covariance [math]\displaystyle{ \Sigma = (\sigma_{ij})_{p\times p} }[/math]
- Diagonal structure: [math]\displaystyle{ \sigma_{ij} = 0 }[/math] if [math]\displaystyle{ i \neq j }[/math].
- Compound symmetry: [math]\displaystyle{ \sigma_{ij} = \rho }[/math] if [math]\displaystyle{ i \neq j }[/math].
- First-order autoregressive AR(1) structure: [math]\displaystyle{ \sigma_{ij} = \rho^{|i - j|} }[/math].
rho <- .8 p <- 5 blockMat <- rho ^ abs(matrix(1:p, p, p, byrow=T) - matrix(1:p, p, p))
- Banded matrix: [math]\displaystyle{ \sigma_{ii}=1, \sigma_{i,i+1}=\sigma_{i+1,i} \neq 0, \sigma_{i,i+2}=\sigma_{i+2,i} \neq 0 }[/math] and [math]\displaystyle{ \sigma_{ij}=0 }[/math] for [math]\displaystyle{ |i-j| \ge 3 }[/math].
- Spatial Power
- Unstructured Covariance
- Toeplitz structure
To create blocks of correlation matrix, use the "%x%" operator. See kronecker().
covMat <- diag(n.blocks) %x% blockMat
Counter/Special Examples
Suppose X is a normally-distributed random variable with zero mean. Let Y = X^2. Clearly X and Y are not independent: if you know X, you also know Y. And if you know Y, you know the absolute value of X.
The covariance of X and Y is
Cov(X,Y) = E(XY) - E(X)E(Y) = E(X^3) - 0*E(Y) = E(X^3)
= 0,
because the distribution of X is symmetric around zero. Thus the correlation r(X,Y) = Cov(X,Y)/Sqrt[Var(X)Var(Y)] = 0, and we have a situation where the variables are not independent, yet have (linear) correlation r(X,Y) = 0.
This example shows how a linear correlation coefficient does not encapsulate anything about the quadratic dependence of Y upon X.
Spearman vs Pearson correlation
Pearson benchmarks linear relationship, Spearman benchmarks monotonic relationship. https://stats.stackexchange.com/questions/8071/how-to-choose-between-pearson-and-spearman-correlation
x=(1:100); y=exp(x); cor(x,y, method='spearman') # 1 cor(x,y, method='pearson') # .25
Spearman vs Wilcoxon
By this post
- Wilcoxon used to compare categorical versus non-normal continuous variable
- Spearman's rho used to compare two continuous (including ordinal) variables that one or both aren't normally distributed
Anscombe quartet
Four datasets have almost same properties: same mean in X, same mean in Y, same variance in X, (almost) same variance in Y, same correlation in X and Y, same linear regression.
The real meaning of spurious correlations
https://nsaunders.wordpress.com/2017/02/03/the-real-meaning-of-spurious-correlations/
library(ggplot2)
set.seed(123)
spurious_data <- data.frame(x = rnorm(500, 10, 1),
y = rnorm(500, 10, 1),
z = rnorm(500, 30, 3))
cor(spurious_data$x, spurious_data$y)
# [1] -0.05943856
spurious_data %>% ggplot(aes(x, y)) + geom_point(alpha = 0.3) +
theme_bw() + labs(title = "Plot of y versus x for 500 observations with N(10, 1)")
cor(spurious_data$x / spurious_data$z, spurious_data$y / spurious_data$z)
# [1] 0.4517972
spurious_data %>% ggplot(aes(x/z, y/z)) + geom_point(aes(color = z), alpha = 0.5) +
theme_bw() + geom_smooth(method = "lm") +
scale_color_gradientn(colours = c("red", "white", "blue")) +
labs(title = "Plot of y/z versus x/z for 500 observations with x,y N(10, 1); z N(30, 3)")
spurious_data$z <- rnorm(500, 30, 6)
cor(spurious_data$x / spurious_data$z, spurious_data$y / spurious_data$z)
# [1] 0.8424597
spurious_data %>% ggplot(aes(x/z, y/z)) + geom_point(aes(color = z), alpha = 0.5) +
theme_bw() + geom_smooth(method = "lm") +
scale_color_gradientn(colours = c("red", "white", "blue")) +
labs(title = "Plot of y/z versus x/z for 500 observations with x,y N(10, 1); z N(30, 6)")
Time series
Structural change
Structural Changes in Global Warming
Dictionary
- Prognosis is the probability that an event or diagnosis will result in a particular outcome.
Data
Eleven quick tips for finding research data
http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1006038