Statistics
Statisticians
- Karl Pearson (1857-1936): chi-square, p-value, PCA
- William Sealy Gosset (1876-1937): Student's t
- Ronald Fisher (1890-1962): ANOVA
- Egon Pearson (1895-1980): son of Karl Pearson
- Jerzy Neyman (1894-1981): type 1 error
Statistics for biologists
http://www.nature.com/collections/qghhqm
Transform sample values to their percentiles
https://stackoverflow.com/questions/21219447/calculating-percentile-of-dataset-column
set.seed(1234) x <- rnorm(10) x # [1] -1.2070657 0.2774292 1.0844412 -2.3456977 0.4291247 0.5060559 # [7] -0.5747400 -0.5466319 -0.5644520 -0.8900378 ecdf(x)(x) # [1] 0.2 0.7 1.0 0.1 0.8 0.9 0.4 0.6 0.5 0.3 rank(x) # [1] 2 7 10 1 8 9 4 6 5 3
Box(Box and whisker) plot in R
See
- https://en.wikipedia.org/wiki/Box_plot
- https://owi.usgs.gov/blog/boxplots/ (ggplot2 is used)
- https://flowingdata.com/2008/02/15/how-to-read-and-use-a-box-and-whisker-plot/
- Quartile from Wikipedia. The quartiles returned from R are the same as the method defined by Method 2 described in Wikipedia.
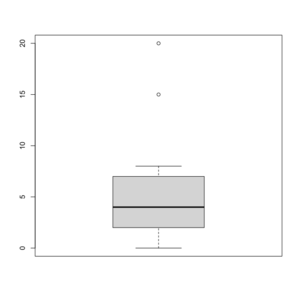
An example for a graphical explanation.
> x=c(0,4,15, 1, 6, 3, 20, 5, 8, 1, 3)
> summary(x)
Min. 1st Qu. Median Mean 3rd Qu. Max.
0 2 4 6 7 20
> sort(x)
[1] 0 1 1 3 3 4 5 6 8 15 20
> boxplot(x, col = 'grey')
# https://en.wikipedia.org/wiki/Quartile#Example_1
> summary(c(6, 7, 15, 36, 39, 40, 41, 42, 43, 47, 49))
Min. 1st Qu. Median Mean 3rd Qu. Max.
6.00 25.50 40.00 33.18 42.50 49.00
- The lower and upper edges of box is determined by the first and 3rd quartiles (2 and 7 in the above example).
- 2 = median(c(0, 1, 1, 3, 3, 4)) = (1+3)/2
- 7 = median(c(4, 5, 6, 8, 15, 20)) = (6+8)/2
- IQR = 7 - 2 = 5
- The thick dark horizon line is the median (4 in the example).
- Outliers are defined by (the empty circles in the plot)
- Observations larger than 3rd quartile + 1.5 * IQR (7+1.5*5=14.5) and
- smaller than 1st quartile - 1.5 * IQR (2-1.5*5=-5.5).
- Note that the cutoffs are not shown in the Box plot.
- Whisker (defined using the cutoffs used to define outliers)
- Upper whisker is defined by the largest "data" below 3rd quartile + 1.5 * IQR (8 in this example), and
- Lower whisker is defined by the smallest "data" greater than 1st quartile - 1.5 * IQR (0 in this example).
- See another example below where we can see the whiskers fall on observations.
Note the wikipedia lists several possible definitions of a whisker. R uses the 2nd method (Tukey boxplot) to define whiskers.
Create boxplots from a list object
Normally we use a vector to create a single boxplot or a formula on a data to create boxplots.
But we can also use split() to create a list and then make boxplots.
Dot-box plot
- http://civilstat.com/2012/09/the-grammar-of-graphics-notes-on-first-reading/
- http://www.r-graph-gallery.com/89-box-and-scatter-plot-with-ggplot2/
- http://www.sthda.com/english/wiki/ggplot2-box-plot-quick-start-guide-r-software-and-data-visualization
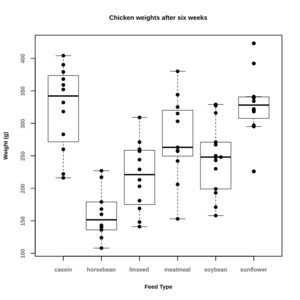
- Graphs in R – Overlaying Data Summaries in Dotplots. Note that for some reason, the boxplot will cover the dots when we save the plot to an svg or a png file. So an alternative solution is to change the order
par(cex.main=0.9,cex.lab=0.8,font.lab=2,cex.axis=0.8,font.axis=2,col.axis="grey50") boxplot(weight ~ feed, data = chickwts, range=0, whisklty = 0, staplelty = 0) par(new = TRUE) stripchart(weight ~ feed, data = chickwts, xlim=c(0.5,6.5), vertical=TRUE, method="stack", offset=0.8, pch=19, main = "Chicken weights after six weeks", xlab = "Feed Type", ylab = "Weight (g)")
Other boxplots
stem and leaf plot
stem(). See R Tutorial.
Note that stem plot is useful when there are outliers.
> stem(x) The decimal point is 10 digit(s) to the right of the | 0 | 00000000000000000000000000000000000000000000000000000000000000000000+419 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 9 > max(x) [1] 129243100275 > max(x)/1e10 [1] 12.92431 > stem(y) The decimal point is at the | 0 | 014478 1 | 0 2 | 1 3 | 9 4 | 8 > y [1] 3.8667356428 0.0001762708 0.7993462430 0.4181079732 0.9541728562 [6] 4.7791262101 0.6899313108 2.1381289177 0.0541736818 0.3868776083 > set.seed(1234) > z <- rnorm(10)*10 > z [1] -12.070657 2.774292 10.844412 -23.456977 4.291247 5.060559 [7] -5.747400 -5.466319 -5.644520 -8.900378 > stem(z) The decimal point is 1 digit(s) to the right of the | -2 | 3 -1 | 2 -0 | 9665 0 | 345 1 | 1
Box-Cox transformation
the Holy Trinity (LRT, Wald, Score tests)
- https://en.wikipedia.org/wiki/Likelihood_function which includes profile likelihood and partial likelihood
- Review of the likelihood theory
- The “Three Plus One” Likelihood-Based Test Statistics: Unified Geometrical and Graphical Interpretations
- Variable selection – A review and recommendations for the practicing statistician by Heinze et al 2018.
- Score test is step-up. Score test is typically used in forward steps to screen covariates currently not included in a model for their ability to improve model.
- Wald test is step-down. Wald test starts at the full model. It evaluate the significance of a variable by comparing the ratio of its estimate and its standard error with an appropriate t distribution (for linear models) or standard normal distribution (for logistic or Cox regression).
- Likelihood ratio tests provide the best control over nuisance parameters by maximizing the likelihood over them both in H0 model and H1 model. In particular, if several coefficients are being tested simultaneously, LRTs for model comparison are preferred over Wald or score tests.
- R packages
- lmtest package, waldtest() and lrtest().
Don't invert that matrix
- http://www.johndcook.com/blog/2010/01/19/dont-invert-that-matrix/
- http://civilstat.com/2015/07/dont-invert-that-matrix-why-and-how/
Different matrix decompositions/factorizations
- QR decomposition, qr()
- LU decomposition, lu() from the 'Matrix' package
- Cholesky decomposition, chol()
- Singular value decomposition, svd()
set.seed(1234) x <- matrix(rnorm(10*2), nr= 10) cmat <- cov(x); cmat # [,1] [,2] # [1,] 0.9915928 -0.1862983 # [2,] -0.1862983 1.1392095 # cholesky decom d1 <- chol(cmat) t(d1) %*% d1 # equal to cmat d1 # upper triangle # [,1] [,2] # [1,] 0.9957875 -0.1870864 # [2,] 0.0000000 1.0508131 # svd d2 <- svd(cmat) d2$u %*% diag(d2$d) %*% t(d2$v) # equal to cmat d2$u %*% diag(sqrt(d2$d)) # [,1] [,2] # [1,] -0.6322816 0.7692937 # [2,] 0.9305953 0.5226872
Linear Regression
Regression Models for Data Science in R by Brian Caffo
Comic https://xkcd.com/1725/
Different models (in R)
http://www.quantide.com/raccoon-ch-1-introduction-to-linear-models-with-r/
dummy.coef.lm() in R
Extracts coefficients in terms of the original levels of the coefficients rather than the coded variables.
Contrasts in linear regression
- Page 147 of Modern Applied Statistics with S (4th ed)
- https://biologyforfun.wordpress.com/2015/01/13/using-and-interpreting-different-contrasts-in-linear-models-in-r/ This explains the meanings of 'treatment', 'helmert' and 'sum' contrasts.
- A (sort of) Complete Guide to Contrasts in R by Rose Maier
mat ## constant NLvMH NvL MvH ## [1,] 1 -0.5 0.5 0.0 ## [2,] 1 -0.5 -0.5 0.0 ## [3,] 1 0.5 0.0 0.5 ## [4,] 1 0.5 0.0 -0.5 mat <- mat[ , -1] model7 <- lm(y ~ dose, data=data, contrasts=list(dose=mat) ) summary(model7) ## Coefficients: ## Estimate Std. Error t value Pr(>|t|) ## (Intercept) 118.578 1.076 110.187 < 2e-16 *** ## doseNLvMH 3.179 2.152 1.477 0.14215 ## doseNvL -8.723 3.044 -2.866 0.00489 ** ## doseMvH 13.232 3.044 4.347 2.84e-05 *** # double check your contrasts attributes(model7$qr$qr)$contrasts ## $dose ## NLvMH NvL MvH ## None -0.5 0.5 0.0 ## Low -0.5 -0.5 0.0 ## Med 0.5 0.0 0.5 ## High 0.5 0.0 -0.5 library(dplyr) dose.means <- summarize(group_by(data, dose), y.mean=mean(y)) dose.means ## Source: local data frame [4 x 2] ## ## dose y.mean ## 1 None 112.6267 ## 2 Low 121.3500 ## 3 Med 126.7839 ## 4 High 113.5517 # The coefficient estimate for the first contrast (3.18) equals the average of # the last two groups (126.78 + 113.55 /2 = 120.17) minus the average of # the first two groups (112.63 + 121.35 /2 = 116.99).
Multicollinearity
- Multicollinearity in R
- alias: Find Aliases (Dependencies) In A Model
> op <- options(contrasts = c("contr.helmert", "contr.poly"))
> npk.aov <- aov(yield ~ block + N*P*K, npk)
> alias(npk.aov)
Model :
yield ~ block + N * P * K
Complete :
(Intercept) block1 block2 block3 block4 block5 N1 P1 K1 N1:P1 N1:K1 P1:K1
N1:P1:K1 0 1 1/3 1/6 -3/10 -1/5 0 0 0 0 0 0
> options(op)
Exposure
https://en.mimi.hu/mathematics/exposure_variable.html
Independent variable = predictor = explanatory = exposure variable
Confounders, confounding
- https://en.wikipedia.org/wiki/Confounding
- A method for controlling complex confounding effects in the detection of adverse drug reactions using electronic health records. It provides a rule to identify a confounder.
- http://anythingbutrbitrary.blogspot.com/2016/01/how-to-create-confounders-with.html (R example)
- Logistic Regression: Confounding and Colinearity
- Identifying a confounder
- Is it possible to have a variable that acts as both an effect modifier and a confounder?
- Which test to use to check if a possible confounder impacts a 0 / 1 result?
- Addressing confounding artifacts in reconstruction of gene co-expression networks Parsana 2019
Confidence interval vs prediction interval
Confidence intervals tell you about how well you have determined the mean E(Y). Prediction intervals tell you where you can expect to see the next data point sampled. That is, CI is computed using Var(E(Y|X)) and PI is computed using Var(E(Y|X) + e).
- http://www.graphpad.com/support/faqid/1506/
- http://en.wikipedia.org/wiki/Prediction_interval
- http://robjhyndman.com/hyndsight/intervals/
- https://stat.duke.edu/courses/Fall13/sta101/slides/unit7lec3H.pdf
- https://datascienceplus.com/prediction-interval-the-wider-sister-of-confidence-interval/
Heteroskedasticity
Dealing with heteroskedasticity; regression with robust standard errors using R
Linear regression with Map Reduce
https://freakonometrics.hypotheses.org/53269
Non- and semi-parametric regression
- Semiparametric Regression in R
- https://socialsciences.mcmaster.ca/jfox/Courses/Oxford-2005/R-nonparametric-regression.html
Splines
- https://en.wikipedia.org/wiki/B-spline
- Cubic and Smoothing Splines in R. bs() is for cubic spline and smooth.spline() is for smoothing spline.
- Can we use B-splines to generate non-linear data?
- How to force passing two data points? (cobs package)
- https://www.rdocumentation.org/packages/cobs/versions/1.3-3/topics/cobs
k-Nearest neighbor regression
- k-NN regression in practice: boundary problem, discontinuities problem.
- Weighted k-NN regression: want weight to be small when distance is large. Common choices - weight = kernel(xi, x)
Kernel regression
- Instead of weighting NN, weight ALL points. Nadaraya-Watson kernel weighted average:
[math]\displaystyle{ \hat{y}_q = \sum c_{qi} y_i/\sum c_{qi} = \frac{\sum \text{Kernel}_\lambda(\text{distance}(x_i, x_q))*y_i}{\sum \text{Kernel}_\lambda(\text{distance}(x_i, x_q))} }[/math].
- Choice of bandwidth [math]\displaystyle{ \lambda }[/math] for bias, variance trade-off. Small [math]\displaystyle{ \lambda }[/math] is over-fitting. Large [math]\displaystyle{ \lambda }[/math] can get an over-smoothed fit. Cross-validation.
- Kernel regression leads to locally constant fit.
- Issues with high dimensions, data scarcity and computational complexity.
Principal component analysis
R source code
> stats:::prcomp.default
function (x, retx = TRUE, center = TRUE, scale. = FALSE, tol = NULL,
...)
{
x <- as.matrix(x)
x <- scale(x, center = center, scale = scale.)
cen <- attr(x, "scaled:center")
sc <- attr(x, "scaled:scale")
if (any(sc == 0))
stop("cannot rescale a constant/zero column to unit variance")
s <- svd(x, nu = 0)
s$d <- s$d/sqrt(max(1, nrow(x) - 1))
if (!is.null(tol)) {
rank <- sum(s$d > (s$d[1L] * tol))
if (rank < ncol(x)) {
s$v <- s$v[, 1L:rank, drop = FALSE]
s$d <- s$d[1L:rank]
}
}
dimnames(s$v) <- list(colnames(x), paste0("PC", seq_len(ncol(s$v))))
r <- list(sdev = s$d, rotation = s$v, center = if (is.null(cen)) FALSE else cen,
scale = if (is.null(sc)) FALSE else sc)
if (retx)
r$x <- x %*% s$v
class(r) <- "prcomp"
r
}
<bytecode: 0x000000003296c7d8>
<environment: namespace:stats>
R example
http://genomicsclass.github.io/book/pages/pca_svd.html
pc <- prcomp(x) group <- as.numeric(tab$Tissue) plot(pc$x[, 1], pc$x[, 2], col = group, main = "PCA", xlab = "PC1", ylab = "PC2")
The meaning of colors can be found by palette().
- black
- red
- green3
- blue
- cyan
- magenta
- yellow
- gray
PCA and SVD
Using the SVD to perform PCA makes much better sense numerically than forming the covariance matrix to begin with, since the formation of [math]\displaystyle{ X X^T }[/math] can cause loss of precision.
http://math.stackexchange.com/questions/3869/what-is-the-intuitive-relationship-between-svd-and-pca
AIC/BIC in estimating the number of components
Related to Factor Analysis
- http://www.aaronschlegel.com/factor-analysis-introduction-principal-component-method-r/.
- http://support.minitab.com/en-us/minitab/17/topic-library/modeling-statistics/multivariate/principal-components-and-factor-analysis/differences-between-pca-and-factor-analysis/
In short,
- In Principal Components Analysis, the components are calculated as linear combinations of the original variables. In Factor Analysis, the original variables are defined as linear combinations of the factors.
- In Principal Components Analysis, the goal is to explain as much of the total variance in the variables as possible. The goal in Factor Analysis is to explain the covariances or correlations between the variables.
- Use Principal Components Analysis to reduce the data into a smaller number of components. Use Factor Analysis to understand what constructs underlie the data.
Calculated by Hand
http://strata.uga.edu/software/pdf/pcaTutorial.pdf
Do not scale your matrix
https://privefl.github.io/blog/(Linear-Algebra)-Do-not-scale-your-matrix/
Visualization
- PCA and Visualization
- Scree plots from the FactoMineR package (based on ggplot2)
What does it do if we choose center=FALSE in prcomp()?
In USArrests data, use center=FALSE gives a better scatter plot of the first 2 PCA components.
x1 = prcomp(USArrests) x2 = prcomp(USArrests, center=F) plot(x1$x[,1], x1$x[,2]) # looks random windows(); plot(x2$x[,1], x2$x[,2]) # looks good in some sense
Relation to Multidimensional scaling/MDS
With no missing data, classical MDS (Euclidean distance metric) is the same as PCA.
Comparisons are here.
Differences are asked/answered on stackexchange.com. The post also answered the question when these two are the same.
isoMDS (Non-metric)
cmdscale (Metric)
Matrix factorization methods
http://joelcadwell.blogspot.com/2015/08/matrix-factorization-comes-in-many.html Review of principal component analysis (PCA), K-means clustering, nonnegative matrix factorization (NMF) and archetypal analysis (AA).
Number of components
Obtaining the number of components from cross validation of principal components regression
Partial Least Squares (PLS)
- https://en.wikipedia.org/wiki/Partial_least_squares_regression. The general underlying model of multivariate PLS is
- [math]\displaystyle{ X = T P^\mathrm{T} + E }[/math]
- [math]\displaystyle{ Y = U Q^\mathrm{T} + F }[/math]
where X is an [math]\displaystyle{ n \times m }[/math] matrix of predictors, Y is an [math]\displaystyle{ n \times p }[/math] matrix of responses; T and U are [math]\displaystyle{ n \times l }[/math] matrices that are, respectively, projections of X (the X score, component or factor matrix) and projections of Y (the Y scores); P and Q are, respectively, [math]\displaystyle{ m \times l }[/math] and [math]\displaystyle{ p \times l }[/math] orthogonal loading matrices; and matrices E and F are the error terms, assumed to be independent and identically distributed random normal variables. The decompositions of X and Y are made so as to maximise the covariance between T and U (projection matrices).
- Supervised vs. Unsupervised Learning: Exploring Brexit with PLS and PCA
- pls R package
- plsRcox R package (archived). See here for the installation.
PLS, PCR (principal components regression) and ridge regression tend to behave similarly. Ridge regression may be preferred because it shrinks smoothly, rather than in discrete steps.
High dimension
Partial least squares prediction in high-dimensional regression Cook and Forzani, 2019
Independent component analysis
ICA is another dimensionality reduction method.
ICA vs PCA
ICS vs FA
Correspondence analysis
https://francoishusson.wordpress.com/2017/07/18/multiple-correspondence-analysis-with-factominer/ and the book Exploratory Multivariate Analysis by Example Using R
t-SNE
t-Distributed Stochastic Neighbor Embedding (t-SNE) is a technique for dimensionality reduction that is particularly well suited for the visualization of high-dimensional datasets.
- https://distill.pub/2016/misread-tsne/
- https://lvdmaaten.github.io/tsne/
- Application to ARCHS4
- Visualization of High Dimensional Data using t-SNE with R
- http://blog.thegrandlocus.com/2018/08/a-tutorial-on-t-sne-1
- Quick and easy t-SNE analysis in R
Visualize the random effects
http://www.quantumforest.com/2012/11/more-sense-of-random-effects/
Calibration
- How to determine calibration accuracy/uncertainty of a linear regression?
- Linear Regression and Calibration Curves
- Regression and calibration Shaun Burke
- calibrate package
- The index of prediction accuracy: an intuitive measure useful for evaluating risk prediction models by Kattan and Gerds 2018. The following code demonstrates Figure 2.
# Odds ratio =1 and calibrated model set.seed(666) x = rnorm(1000) z1 = 1 + 0*x pr1 = 1/(1+exp(-z1)) y1 = rbinom(1000,1,pr1) mean(y1) # .724, marginal prevalence of the outcome dat1 <- data.frame(x=x, y=y1) newdat1 <- data.frame(x=rnorm(1000), y=rbinom(1000, 1, pr1)) # Odds ratio =1 and severely miscalibrated model set.seed(666) x = rnorm(1000) z2 = -2 + 0*x pr2 = 1/(1+exp(-z2)) y2 = rbinom(1000,1,pr2) mean(y2) # .12 dat2 <- data.frame(x=x, y=y2) newdat2 <- data.frame(x=rnorm(1000), y=rbinom(1000, 1, pr2)) library(riskRegression) lrfit1 <- glm(y ~ x, data = dat1, family = 'binomial') IPA(lrfit1, newdata = newdat1) # Variable Brier IPA IPA.gain # 1 Null model 0.1984710 0.000000e+00 -0.003160010 # 2 Full model 0.1990982 -3.160010e-03 0.000000000 # 3 x 0.1984800 -4.534668e-05 -0.003114664 1 - 0.1990982/0.1984710 # [1] -0.003160159 lrfit2 <- glm(y ~ x, family = 'binomial') IPA(lrfit2, newdata = newdat1) # Variable Brier IPA IPA.gain # 1 Null model 0.1984710 0.000000 -1.859333763 # 2 Full model 0.5674948 -1.859334 0.000000000 # 3 x 0.5669200 -1.856437 -0.002896299 1 - 0.5674948/0.1984710 # [1] -1.859334
From the simulated data, we see IPA = -3.16e-3 for a calibrated model and IPA = -1.86 for a severely miscalibrated model.
ROC curve and Brier score
- Binary case:
- Y = true positive rate = sensitivity,
- X = false positive rate = 1-specificity
- Area under the curve AUC from the wikipedia: the probability that a classifier will rank a randomly chosen positive instance higher than a randomly chosen negative one (assuming 'positive' ranks higher than 'negative').
- [math]\displaystyle{ A = \int_{\infty}^{-\infty} \mbox{TPR}(T) \mbox{FPR}'(T) \, dT = \int_{-\infty}^{\infty} \int_{-\infty}^{\infty} I(T'\gt T)f_1(T') f_0(T) \, dT' \, dT = P(X_1 \gt X_0) }[/math]
where [math]\displaystyle{ X_1 }[/math] is the score for a positive instance and [math]\displaystyle{ X_0 }[/math] is the score for a negative instance, and [math]\displaystyle{ f_0 }[/math] and [math]\displaystyle{ f_1 }[/math] are probability densities as defined in previous section.
- Interpretation of the AUC. A small toy example (n=12=4+8) was used to calculate the exact probability [math]\displaystyle{ P(X_1 \gt X_0) }[/math] (4*8=32 all combinations).
- It is a discrimination measure which tells us how well we can classify patients in two groups: those with and those without the outcome of interest.
- Since the measure is based on ranks, it is not sensitive to systematic errors in the calibration of the quantitative tests.
- The AUC can be defined as The probability that a randomly selected case will have a higher test result than a randomly selected control.
- Plot of sensitivity/specificity (y-axis) vs cutoff points of the biomarker
- The Mann-Whitney U test statistic (or Wilcoxon or Kruskall-Wallis test statistic) is equivalent to the AUC (Mason, 2002)
- The p-value of the Mann-Whitney U test can thus safely be used to test whether the AUC differs significantly from 0.5 (AUC of an uninformative test).
- Calculate AUC by hand. AUC is equal to the probability that a true positive is scored greater than a true negative.
- How to calculate Area Under the Curve (AUC), or the c-statistic, by hand or by R
- Introduction to the ROCR package. Add threshold labels
- http://freakonometrics.hypotheses.org/9066, http://freakonometrics.hypotheses.org/20002
- Illustrated Guide to ROC and AUC
- ROC Curves in Two Lines of R Code
- Gini and AUC. Gini = 2*AUC-1.
- Generally, an AUC value over 0.7 is indicative of a model that can distinguish between the two outcomes well. An AUC of 0.5 tells us that the model is a random classifier, and it cannot distinguish between the two outcomes.
Survival data
'Survival Model Predictive Accuracy and ROC Curves' by Heagerty & Zheng 2005
- Recall Sensitivity= [math]\displaystyle{ P(\hat{p_i} \gt c | Y_i=1) }[/math], Specificity= [math]\displaystyle{ P(\hat{p}_i \le c | Y_i=0 }[/math]), [math]\displaystyle{ Y_i }[/math] is binary outcomes, [math]\displaystyle{ \hat{p}_i }[/math] is a prediction, [math]\displaystyle{ c }[/math] is a criterion for classifying the prediction as positive ([math]\displaystyle{ \hat{p}_i \gt c }[/math]) or negative ([math]\displaystyle{ \hat{p}_i \le c }[/math]).
- For survival data, we need to use a fixed time/horizon (t) to classify the data as either a case or a control. Following Heagerty and Zheng's definition (Incident/dynamic), Sensitivity(c, t)= [math]\displaystyle{ P(M_i \gt c | T_i = t) }[/math], Specificity= [math]\displaystyle{ P(M_i \le c | T_i \gt 0 }[/math]) where M is a marker value or [math]\displaystyle{ Z^T \beta }[/math]. Here sensitivity measures the expected fraction of subjects with a marker greater than c among the subpopulation of individuals who die at time t, while specificity measures the fraction of subjects with a marker less than or equal to c among those who survive beyond time t.
- The AUC measures the probability that the marker value for a randomly selected case exceeds the marker value for a randomly selected control
- ROC curves are useful for comparing the discriminatory capacity of different potential biomarkers.
Confusion matrix, Sensitivity/Specificity/Accuracy
| Predict | ||||
| 1 | 0 | |||
| True | 1 | TP | FN | Sens=TP/(TP+FN)=Recall FNR=FN/(TP+FN) |
| 0 | FP | TN | Spec=TN/(FP+TN) | |
| PPV=TP/(TP+FP) FDR=FP/(TP+FP) |
NPV=TN/(FN+TN) | N = TP + FP + FN + TN | ||
- Sensitivity = TP / (TP + FN) = Recall
- Specificity = TN / (TN + FP)
- Accuracy = (TP + TN) / N
- False discovery rate FDR = FP / (TP + FP)
- False negative rate FNR = FN / (TP + FN)
- Positive predictive value (PPV) = TP / # positive calls = TP / (TP + FP) = 1 - FDR
- Negative predictive value (NPV) = TN / # negative calls = TN / (FN + TN)
- Prevalence = (TP + FN) / N.
- Note that PPV & NPV can also be computed from sensitivity, specificity, and prevalence:
- PPV is directly proportional to the prevalence of the disease or condition..
- For example, in the extreme case if the prevalence =1, then PPV is always 1.
- [math]\displaystyle{ \text{PPV} = \frac{\text{sensitivity} \times \text{prevalence}}{\text{sensitivity} \times \text{prevalence}+(1-\text{specificity}) \times (1-\text{prevalence})} }[/math]
- [math]\displaystyle{ \text{NPV} = \frac{\text{specificity} \times (1-\text{prevalence})}{(1-\text{sensitivity}) \times \text{prevalence}+\text{specificity} \times (1-\text{prevalence})} }[/math]
Precision recall curve
- Precision and recall
- Y-axis: Precision = tp/(tp + fp) = PPV, large is better
- X-axis: Recall = tp/(tp + fn) = Sensitivity, large is better
- The Relationship Between Precision-Recall and ROC Curves. Remember ROC is defined as
- Y-axis: Sensitivity = tp/(tp + fn) = Recall
- X-axis: 1-Specificity = fp/(fp + tn)
Incidence, Prevalence
https://www.health.ny.gov/diseases/chronic/basicstat.htm
Calculate area under curve by hand (using trapezoid), relation to concordance measure and the Wilcoxon–Mann–Whitney test
- https://stats.stackexchange.com/a/146174
- The meaning and use of the area under a receiver operating characteristic (ROC) curve J A Hanley, B J McNeil 1982
genefilter package and rowpAUCs function
- rowpAUCs function in genefilter package. The aim is to find potential biomarkers whose expression level is able to distinguish between two groups.
# source("http://www.bioconductor.org/biocLite.R")
# biocLite("genefilter")
library(Biobase) # sample.ExpressionSet data
data(sample.ExpressionSet)
library(genefilter)
r2 = rowpAUCs(sample.ExpressionSet, "sex", p=0.1)
plot(r2[1]) # first gene, asking specificity = .9
r2 = rowpAUCs(sample.ExpressionSet, "sex", p=1.0)
plot(r2[1]) # it won't show pAUC
r2 = rowpAUCs(sample.ExpressionSet, "sex", p=.999)
plot(r2[1]) # pAUC is very close to AUC now
Use and Misuse of the Receiver Operating Characteristic Curve in Risk Prediction
http://circ.ahajournals.org/content/115/7/928
Performance evaluation
- Testing for improvement in prediction model performance by Pepe et al 2013.
Some R packages
Comparison of two AUCs
- Statistical Assessments of AUC. This is using the pROC::roc.test function.
NRI (Net reclassification improvement)
Maximum likelihood
Difference of partial likelihood, profile likelihood and marginal likelihood
Generalized Linear Model
Lectures from a course in Simon Fraser University Statistics.
Doing magic and analyzing seasonal time series with GAM (Generalized Additive Model) in R
Quasi Likelihood
Quasi-likelihood is like log-likelihood. The quasi-score function (first derivative of quasi-likelihood function) is the estimating equation.
- Original paper by Peter McCullagh.
- Lecture 20 from SFU.
- U. Washington and another lecture focuses on overdispersion.
- This lecture contains a table of quasi likelihood from common distributions.
Plot
Deviance, stats::deviance() and glmnet::deviance.glmnet() from R
- It is a generalization of the idea of using the sum of squares of residuals (RSS) in ordinary least squares to cases where model-fitting is achieved by maximum likelihood. See What is Deviance? (specifically in CART/rpart) to manually compute deviance and compare it with the returned value of the deviance() function from a linear regression. Summary: deviance() = RSS in linear models.
- https://www.rdocumentation.org/packages/stats/versions/3.4.3/topics/deviance
- Likelihood ratio tests and the deviance http://data.princeton.edu/wws509/notes/a2.pdf#page=6
- Deviance(y,muhat) = 2*(loglik_saturated - loglik_proposed)
- Interpreting Residual and Null Deviance in GLM R
- Null Deviance = 2(LL(Saturated Model) - LL(Null Model)) on df = df_Sat - df_Null. The null deviance shows how well the response variable is predicted by a model that includes only the intercept (grand mean).
- Residual Deviance = 2(LL(Saturated Model) - LL(Proposed Model)) = [math]\displaystyle{ 2(LL(y|y) - LL(\hat{\mu}|y)) }[/math], df = df_Sat - df_Proposed=n-p. ==> deviance() has returned.
- Null deviance > Residual deviance. Null deviance df = n-1. Residual deviance df = n-p.
## an example with offsets from Venables & Ripley (2002, p.189)
utils::data(anorexia, package = "MASS")
anorex.1 <- glm(Postwt ~ Prewt + Treat + offset(Prewt),
family = gaussian, data = anorexia)
summary(anorex.1)
# Call:
# glm(formula = Postwt ~ Prewt + Treat + offset(Prewt), family = gaussian,
# data = anorexia)
#
# Deviance Residuals:
# Min 1Q Median 3Q Max
# -14.1083 -4.2773 -0.5484 5.4838 15.2922
#
# Coefficients:
# Estimate Std. Error t value Pr(>|t|)
# (Intercept) 49.7711 13.3910 3.717 0.000410 ***
# Prewt -0.5655 0.1612 -3.509 0.000803 ***
# TreatCont -4.0971 1.8935 -2.164 0.033999 *
# TreatFT 4.5631 2.1333 2.139 0.036035 *
# ---
# Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#
# (Dispersion parameter for gaussian family taken to be 48.69504)
#
# Null deviance: 4525.4 on 71 degrees of freedom
# Residual deviance: 3311.3 on 68 degrees of freedom
# AIC: 489.97
#
# Number of Fisher Scoring iterations: 2
deviance(anorex.1)
# [1] 3311.263
- In glmnet package. The deviance is defined to be 2*(loglike_sat - loglike), where loglike_sat is the log-likelihood for the saturated model (a model with a free parameter per observation). Null deviance is defined to be 2*(loglike_sat -loglike(Null)); The NULL model refers to the intercept model, except for the Cox, where it is the 0 model. Hence dev.ratio=1-deviance/nulldev, and this deviance method returns (1-dev.ratio)*nulldev.
x=matrix(rnorm(100*2),100,2) y=rnorm(100) fit1=glmnet(x,y) deviance(fit1) # one for each lambda # [1] 98.83277 98.53893 98.29499 98.09246 97.92432 97.78472 97.66883 # [8] 97.57261 97.49273 97.41327 97.29855 97.20332 97.12425 97.05861 # ... # [57] 96.73772 96.73770 fit2 <- glmnet(x, y, lambda=.1) # fix lambda deviance(fit2) # [1] 98.10212 deviance(glm(y ~ x)) # [1] 96.73762 sum(residuals(glm(y ~ x))^2) # [1] 96.73762
Saturated model
- The saturated model always has n parameters where n is the sample size.
- Logistic Regression : How to obtain a saturated model
Simulate data
Density plot
# plot a Weibull distribution with shape and scale func <- function(x) dweibull(x, shape = 1, scale = 3.38) curve(func, .1, 10) func <- function(x) dweibull(x, shape = 1.1, scale = 3.38) curve(func, .1, 10)
The shape parameter plays a role on the shape of the density function and the failure rate.
- Shape <=1: density is convex, not a hat shape.
- Shape =1: failure rate (hazard function) is constant. Exponential distribution.
- Shape >1: failure rate increases with time
Simulate data from a specified density
Signal to noise ratio
- https://en.wikipedia.org/wiki/Signal-to-noise_ratio
- https://stats.stackexchange.com/questions/31158/how-to-simulate-signal-noise-ratio
- [math]\displaystyle{ \frac{\sigma^2_{signal}}{\sigma^2_{noise}} = \frac{Var(f(X))}{Var(e)} }[/math] if Y = f(X) + e
- Page 401 of ESLII (https://web.stanford.edu/~hastie/ElemStatLearn//) 12th print.
Some examples of signal to noise ratio
- ESLII_print12.pdf: .64, 5, 4
- Yuan and Lin 2006: 1.8, 3
- A framework for estimating and testing qualitative interactions with applications to predictive biomarkers Roth, Biostatistics, 2018
Effect size, Cohen's d and volcano plot
- https://en.wikipedia.org/wiki/Effect_size (See also the estimation by the pooled sd)
- [math]\displaystyle{ \theta = \frac{\mu_1 - \mu_2} \sigma, }[/math]
- Effect size, sample size and power from Learning statistics with R: A tutorial for psychology students and other beginners.
- t-statistic and Cohen's d for the case of mean difference between two independent groups
- Cohen’s D for Experimental Planning
- Volcano plot
- Y-axis: -log(p)
- X-axis: log2 fold change OR effect size (Cohen's D). An example from RNA-Seq data.
Multiple comparisons
- If you perform experiments over and over, you's bound to find something. So significance level must be adjusted down when performing multiple hypothesis tests.
- http://www.gs.washington.edu/academics/courses/akey/56008/lecture/lecture10.pdf
- Book 'Multiple Comparison Using R' by Bretz, Hothorn and Westfall, 2011.
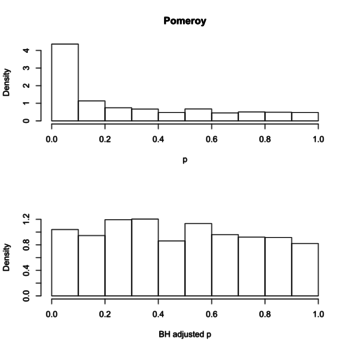
- Plot a histogram of p-values, a post from varianceexplained.org. The anti-conservative histogram (tail on the RHS) is what we have typically seen in e.g. microarray gene expression data.
- Comparison of different ways of multiple-comparison in R.
Take an example, Suppose 550 out of 10,000 genes are significant at .05 level
- P-value < .05 ==> Expect .05*10,000=500 false positives
- False discovery rate < .05 ==> Expect .05*550 =27.5 false positives
- Family wise error rate < .05 ==> The probablity of at least 1 false positive <.05
According to Lifetime Risk of Developing or Dying From Cancer, there is a 39.7% risk of developing a cancer for male during his lifetime (in other words, 1 out of every 2.52 men in US will develop some kind of cancer during his lifetime) and 37.6% for female. So the probability of getting at least one cancer patient in a 3-generation family is 1-.6**3 - .63**3 = 0.95.
False Discovery Rate
- https://en.wikipedia.org/wiki/False_discovery_rate
- Paper Definition by Benjamini and Hochberg in JRSS B 1995.
- A comic
- P-value vs false discovery rate vs family wise error rate. See 10 statistics tip or Statistics for Genomics (140.688) from Jeff Leek. Suppose 550 out of 10,000 genes are significant at .05 level
- P-value < .05 implies expecting .05*10000 = 500 false positives
- False discovery rate < .05 implies expecting .05*550 = 27.5 false positives
- Family wise error rate (P (# of false positives ≥ 1)) < .05. See Understanding Family-Wise Error Rate
- Statistical significance for genomewide studies by Storey and Tibshirani.
- What’s the probability that a significant p-value indicates a true effect?
- http://onetipperday.sterding.com/2015/12/my-note-on-multiple-testing.html
- A practical guide to methods controlling false discoveries in computational biology by Korthauer, et al 2018, BMC Genome Biology 2019
- onlineFDR: an R package to control the false discovery rate for growing data repositories
Suppose [math]\displaystyle{ p_1 \leq p_2 \leq ... \leq p_n }[/math]. Then
- [math]\displaystyle{ \text{FDR}_i = \text{min}(1, n* p_i/i) }[/math].
So if the number of tests ([math]\displaystyle{ n }[/math]) is large and/or the original p value ([math]\displaystyle{ p_i }[/math]) is large, then FDR can hit the value 1.
However, the simple formula above does not guarantee the monotonicity property from the FDR. So the calculation in R is more complicated. See How Does R Calculate the False Discovery Rate.
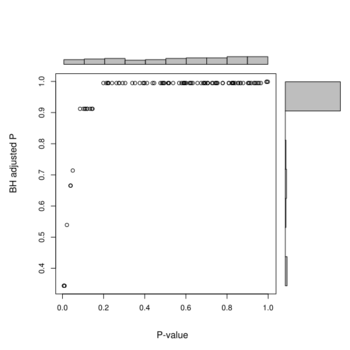
Below is the histograms of p-values and FDR (BH adjusted) from a real data (Pomeroy in BRB-ArrayTools).
And the next is a scatterplot w/ histograms on the margins from a null data.
q-value
q-value is defined as the minimum FDR that can be attained when calling that feature significant (i.e., expected proportion of false positives incurred when calling that feature significant).
If gene X has a q-value of 0.013 it means that 1.3% of genes that show p-values at least as small as gene X are false positives.
SAM/Significance Analysis of Microarrays
The percentile option is used to define the number of falsely called genes based on 'B' permutations. If we use the 90-th percentile, the number of significant genes will be less than if we use the 50-th percentile/median.
In BRCA dataset, using the 90-th percentile will get 29 genes vs 183 genes if we use median.
Multivariate permutation test
In BRCA dataset, using 80% confidence gives 116 genes vs 237 genes if we use 50% confidence (assuming maximum proportion of false discoveries is 10%). The method is published on EL Korn, JF Troendle, LM McShane and R Simon, Controlling the number of false discoveries: Application to high dimensional genomic data, Journal of Statistical Planning and Inference, vol 124, 379-398 (2004).
String Permutations Algorithm
Solving the Empirical Bayes Normal Means Problem with Correlated Noise Sun 2018
The package cashr and the source code of the paper
Bayes
Bayes factor
Empirical Bayes method
- http://en.wikipedia.org/wiki/Empirical_Bayes_method
- Introduction to Empirical Bayes: Examples from Baseball Statistics
Naive Bayes classifier
Understanding Naïve Bayes Classifier Using R
MCMC
Speeding up Metropolis-Hastings with Rcpp
offset() function
- An offset is a term to be added to a linear predictor, such as in a generalised linear model, with known coefficient 1 rather than an estimated coefficient.
- https://www.rdocumentation.org/packages/stats/versions/3.5.0/topics/offset
Offset in Poisson regression
- http://rfunction.com/archives/223
- https://stats.stackexchange.com/questions/11182/when-to-use-an-offset-in-a-poisson-regression
- We need to model rates instead of counts
- More generally, you use offsets because the units of observation are different in some dimension (different populations, different geographic sizes) and the outcome is proportional to that dimension.
An example from here
Y <- c(15, 7, 36, 4, 16, 12, 41, 15) N <- c(4949, 3534, 12210, 344, 6178, 4883, 11256, 7125) x1 <- c(-0.1, 0, 0.2, 0, 1, 1.1, 1.1, 1) x2 <- c(2.2, 1.5, 4.5, 7.2, 4.5, 3.2, 9.1, 5.2) glm(Y ~ offset(log(N)) + (x1 + x2), family=poisson) # two variables # Coefficients: # (Intercept) x1 x2 # -6.172 -0.380 0.109 # # Degrees of Freedom: 7 Total (i.e. Null); 5 Residual # Null Deviance: 10.56 # Residual Deviance: 4.559 AIC: 46.69 glm(Y ~ offset(log(N)) + I(x1+x2), family=poisson) # one variable # Coefficients: # (Intercept) I(x1 + x2) # -6.12652 0.04746 # # Degrees of Freedom: 7 Total (i.e. Null); 6 Residual # Null Deviance: 10.56 # Residual Deviance: 8.001 AIC: 48.13
Offset in Cox regression
An example from biospear::PCAlasso()
coxph(Surv(time, status) ~ offset(off.All), data = data) # Call: coxph(formula = Surv(time, status) ~ offset(off.All), data = data) # # Null model # log likelihood= -2391.736 # n= 500 # versus without using offset() coxph(Surv(time, status) ~ off.All, data = data) # Call: # coxph(formula = Surv(time, status) ~ off.All, data = data) # # coef exp(coef) se(coef) z p # off.All 0.485 1.624 0.658 0.74 0.46 # # Likelihood ratio test=0.54 on 1 df, p=0.5 # n= 500, number of events= 438 coxph(Surv(time, status) ~ off.All, data = data)$loglik # [1] -2391.702 -2391.430 # initial coef estimate, final coef
Offset in linear regression
- https://www.rdocumentation.org/packages/stats/versions/3.5.1/topics/lm
- https://stackoverflow.com/questions/16920628/use-of-offset-in-lm-regression-r
Overdispersion
https://en.wikipedia.org/wiki/Overdispersion
Var(Y) = phi * E(Y). If phi > 1, then it is overdispersion relative to Poisson. If phi <1, we have under-dispersion (rare).
Heterogeneity
The Poisson model fit is not good; residual deviance/df >> 1. The lack of fit maybe due to missing data, covariates or overdispersion.
Subjects within each covariate combination still differ greatly.
- https://onlinecourses.science.psu.edu/stat504/node/169.
- https://onlinecourses.science.psu.edu/stat504/node/162
Consider Quasi-Poisson or negative binomial.
Test of overdispersion or underdispersion in Poisson models
Negative Binomial
The mean of the Poisson distribution can itself be thought of as a random variable drawn from the gamma distribution thereby introducing an additional free parameter.
Binomial
- Generating and modeling over-dispersed binomial data
- simstudy package. The final data sets can represent data from randomized control trials, repeated measure (longitudinal) designs, and cluster randomized trials. Missingness can be generated using various mechanisms (MCAR, MAR, NMAR).
Count data
Zero counts
Bias
Bias in Small-Sample Inference With Count-Data Models Blackburn 2019
Survival data
- A Package for Survival Analysis in S by Terry M. Therneau, 1999
- https://web.stanford.edu/~lutian/coursepdf/stat331.HTML and https://web.stanford.edu/~lutian/coursepdf/ (3 types of tests).
- http://www.stat.columbia.edu/~madigan/W2025/notes/survival.pdf.
- How to manually compute the KM curve and by R
- Estimation of parametric survival function from joint likelihood in theory and R.
- http://data.princeton.edu/wws509/notes/c7s1.html
- http://data.princeton.edu/pop509/ParametricSurvival.pdf Parametric survival models with covariates (logT = alpha + sigma W) p8
- Weibull p2 where T ~ Weibull and W ~ Extreme value.
- Gamma p3 where T ~ Gamma and W ~ Generalized extreme value
- Generalized gamma p4,
- log normal p4 where T ~ lognormal and W ~ N(0,1)
- log logistic p4 where T ~ log logistic and W ~ standard logistic distribution.
- http://www.math.ucsd.edu/~rxu/math284/ (good cover) Wald test
- http://www.stats.ox.ac.uk/~mlunn/
- https://www.openintro.org/download.php?file=survival_analysis_in_R&referrer=/stat/surv.php
- https://cran.r-project.org/web/packages/survival/vignettes/timedep.pdf
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1065034/
- Survival Analysis with R from rviews.rstudio.com
- Survival Analysis with R from bioconnector.og.
Censoring
Sample schemes of incomplete data
- Type I censoring: the censoring time is fixed
- Type II censoring
- Random censoring
- Right censoring
- Left censoring
- Interval censoring
- Truncation
The most common is called right censoring and occurs when a participant does not have the event of interest during the study and thus their last observed follow-up time is less than their time to event. This can occur when a participant drops out before the study ends or when a participant is event free at the end of the observation period.
Definitions of common terms in survival analysis
- Event: Death, disease occurrence, disease recurrence, recovery, or other experience of interest
- Time: The time from the beginning of an observation period (such as surgery or beginning treatment) to (i) an event, or (ii) end of the study, or (iii) loss of contact or withdrawal from the study.
- Censoring / Censored observation: If a subject does not have an event during the observation time, they are described as censored. The subject is censored in the sense that nothing is observed or known about that subject after the time of censoring. A censored subject may or may not have an event after the end of observation time.
In R, "status" should be called event status. status = 1 means event occurred. status = 0 means no event (censored). Sometimes the status variable has more than 2 states. We can uses "status != 0" to replace "status" in Surv() function.
- status=0/1/2 for censored, transplant and dead in survival::pbc data.
- status=0/1/2 for censored, relapse and dead in randomForestSRC::follic data.
How to explore survival data
https://en.wikipedia.org/wiki/Survival_analysis#Survival_analysis_in_R
- Create graph of length of time that each subject was in the study
<syntaxhighlight lang='rsplus'>