File list
Appearance
This special page shows all uploaded files.
| Date | Name | Thumbnail | Size | User | Description | Versions |
|---|---|---|---|---|---|---|
| 06:08, 27 August 2022 | FgseaPlotTop.png (file) | 15 KB | Brb (talk | contribs) | 1 | ||
| 13:17, 29 August 2022 | Rbiomirgs volcano.png (file) |  |
66 KB | Brb (talk | contribs) | 1 | |
| 13:17, 29 August 2022 | Rbiomirgs bar.png (file) |  |
92 KB | Brb (talk | contribs) | 1 | |
| 13:19, 29 August 2022 | Rbiomirgs barall.png (file) | 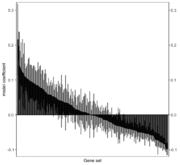 |
35 KB | Brb (talk | contribs) | 1 | |
| 09:08, 30 August 2022 | ScaleFillViridisDiscrete.png (file) | 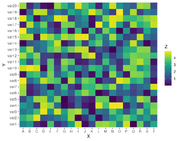 |
22 KB | Brb (talk | contribs) | See https://r-graph-gallery.com/79-levelplot-with-ggplot2.html <pre> library(ggplot2) # library(hrbrthemes) # Dummy data x <- LETTERS[1:20] y <- paste0("var", seq(1,20)) data <- expand.grid(X=x, Y=y) data$Z <- runif(400, 0, 5) library(viridis) ggplot(data, aes(X, Y, fill= Z)) + geom_tile() + scale_fill_viridis(discrete=FALSE) </pre> | 1 |
| 09:31, 30 August 2022 | ViridisDefault.png (file) | 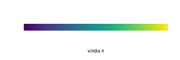 |
2 KB | Brb (talk | contribs) | <pre> library(viridis) n = 200 image( 1:n, 1, as.matrix(1:n), col = viridis(n, option = "D"), xlab = "viridis n", ylab = "", xaxt = "n", yaxt = "n", bty = "n" ) </pre> | 1 |
| 10:50, 30 August 2022 | Geomcolviridis.png (file) |  |
26 KB | Brb (talk | contribs) | Modify the example from https://datavizpyr.com/re-ordering-bars-in-barplot-in-r/ to allow filled colors and facet. <pre> library(tidyverse) library(gapminder) library(viridis) theme_set(theme_bw(base_size=16)) pop_df <- gapminder %>% filter(year==2007)%>% group_by(continent) %>% summarize(pop_in_millions=sum(pop)/1e06) pop_df2 <- tibble(class=rbinom(nrow(pop_df), 1, .5), pop_df) pop_df2 <- pop_df2 |> mutate(pop_in_millions = pop_in_millions-1900) pop_df2 %>% ggplot(aes... | 1 |
| 10:06, 6 October 2022 | Barplot ggplot2.png (file) | 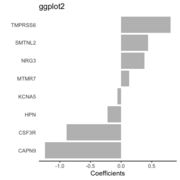 |
16 KB | Brb (talk | contribs) | 1 | |
| 10:11, 6 October 2022 | Barplot base.png (file) | 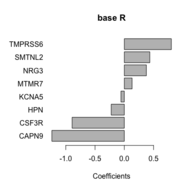 |
20 KB | Brb (talk | contribs) | 2 | |
| 13:12, 6 October 2022 | Greedypairs.png (file) | 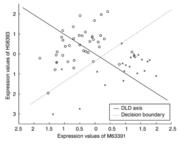 |
29 KB | Brb (talk | contribs) | 1 | |
| 15:39, 11 October 2022 | Cvglmnetplot.png (file) |  |
34 KB | Brb (talk | contribs) | <pre> n <- 100 set.seed(1) x1 <- rnorm(n) e <- rnorm(n)*.01 y <- x1 + e x4 <- x fit <- cv.glmnet(x=cbind(x1, x4, matrix(rnorm(n*10), nr=n)), y=y) plot (fit) </pre> | 1 |
| 10:03, 12 October 2022 | Foldchangefilter.png (file) | 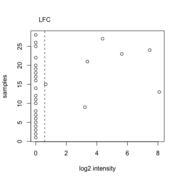 |
22 KB | Brb (talk | contribs) | <pre> LFC <- log2(1.5) x <- c(0, 0, 0, 0, 0, 0, 0, 0, 3.22, 0, 0, 0, 8.09, 0, 0.65, 0, 0, 0, 0, 0, 3.38, 0, 5.63, 7.46, 0, 0, 4.38, 0) plot(x, y = 1:28, xlab="log2 intensity", ylab="samples") abline(v=LFC, lty="dashed") axis(side=3,at=LFC, labels="LFC", tick=FALSE, line=0) </pre> | 1 |
| 16:26, 19 October 2022 | NC better kNN.png (file) | 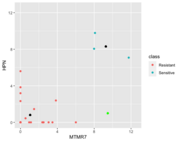 |
19 KB | Brb (talk | contribs) | The green color is a new observation (Sensitive). By using the kNN method, it will be assigned to Resistant b/c it is closer to the Resistant group. However, using the NC, the distance of it to the Resistant group centroid is 8.42 which is larger than the distance of it to the Sensitive groups centroid 7.31. So NC classified it to Sensitive. Color annotation: green=LOO obs, black=centroid in each class. | 1 |
| 09:48, 20 October 2022 | 1NN better NC.png (file) | 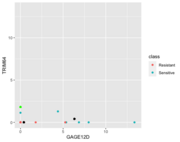 |
18 KB | Brb (talk | contribs) | 1 | |
| 08:46, 27 October 2022 | HC single.png (file) | 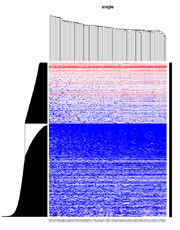 |
133 KB | Brb (talk | contribs) | 1 | |
| 14:15, 27 October 2022 | DHdialog1.png (file) | 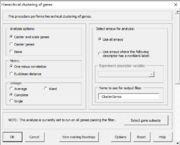 |
21 KB | Brb (talk | contribs) | 1 | |
| 14:15, 27 October 2022 | DHdialog2.png (file) | 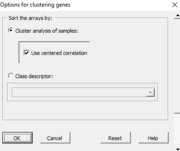 |
6 KB | Brb (talk | contribs) | 1 | |
| 14:39, 29 October 2022 | FgseaPlotSmallm.png (file) | 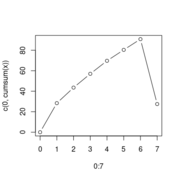 |
7 KB | Brb (talk | contribs) | 1 | |
| 14:55, 29 October 2022 | ExampleRanks.png (file) | 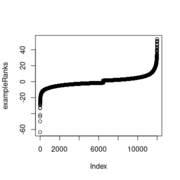 |
6 KB | Brb (talk | contribs) | <pre> plot(exampleRanks) </pre> | 1 |
| 17:00, 5 December 2022 | GpartedinfoSanDisk.png (file) |  |
46 KB | Brb (talk | contribs) | 1 | |
| 13:39, 15 December 2022 | Geom bar1.png (file) | 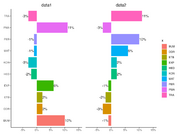 |
85 KB | Brb (talk | contribs) | <pre> library(ggplot2) library(scales) library(patchwork) dtf <- data.frame(x = c("ETB", "PMA", "PER", "KON", "TRA", "DDR", "BUM", "MAT", "HED", "EXP"), y = c(.02, .11, -.01, -.03, -.03, .02, .1, -.01, -.02, 0.06)) set.seed(1) dtf2 <- data.frame(x = dtf[, 1], y = sample(dtf[, 2])) g1 <- ggplot(dtf, aes(x, y)) + geom_bar(stat = "identity", aes(fill = x)) + geom_text(aes(label = paste0(y * 100, "%"), hjust = ifelse(y >= 0,... | 1 |
| 13:41, 15 December 2022 | Geom bar2.png (file) |  |
71 KB | Brb (talk | contribs) | <pre> library(ggplot2) library(scales) library(patchwork) dtf <- data.frame(x = c("ETB", "PMA", "PER", "KON", "TRA", "DDR", "BUM", "MAT", "HED", "EXP"), y = c(.02, .11, -.01, -.03, -.03, .02, .1, -.01, -.02, 0.06)) set.seed(1) dtf2 <- data.frame(x = dtf[, 1], y = sample(dtf[, 2])) g1 <- ggplot(dtf, aes(x, y)) + geom_bar(stat = "identity", fill = "#F8767D") + geom_text(aes(label = paste0(y * 100, "%"), hjust = ifelse(y >= 0, 0, 1))) +... | 1 |
| 14:08, 15 December 2022 | Geom bar3.png (file) | 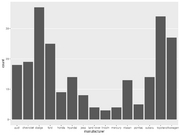 |
44 KB | Brb (talk | contribs) | <pre> ggplot(mpg, aes(x=manufacturer)) + geom_bar() + theme(panel.grid.major.x = element_blank(), panel.grid.minor = element_blank()) </pre> | 1 |
| 14:14, 15 December 2022 | Geom bar4.png (file) | 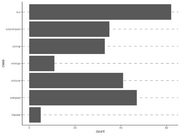 |
51 KB | Brb (talk | contribs) | <pre> ggplot(mpg, aes(x = class)) + geom_vline(xintercept = mpg$class, color = "grey", linetype = "dashed", size = 1) + geom_bar() + theme_classic() + coord_flip() </pre> | 1 |
| 14:40, 15 December 2022 | Geom smooth ex.png (file) | 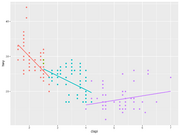 |
110 KB | Brb (talk | contribs) | <pre> library(dplyr) #group the data by cyl and create the plots mpg %>% group_by(cyl) %>% ggplot(aes(x=displ, y=hwy, color=factor(cyl))) + geom_point() + geom_smooth(method = "lm", se = FALSE) + theme(legend.position="none") </pre> | 1 |
| 19:07, 25 December 2022 | DisableDropbox4pm.png (file) |  |
246 KB | Brb (talk | contribs) | 1 | |
| 13:58, 31 December 2022 | Filebrowser.png (file) |  |
86 KB | Brb (talk | contribs) | 1 | |
| 11:37, 6 January 2023 | Fgsea3plots.png (file) |  |
68 KB | Brb (talk | contribs) | <pre> par(mfrow=c(1,3)) with(fgseaRes, plot(abs(ES), pval)) with(fgseaRes, plot(abs(NES), pval)) with(fgseaRes, plot(ES, NES)) </pre> | 1 |
| 20:37, 7 January 2023 | ComplexHeatmap2.png (file) | 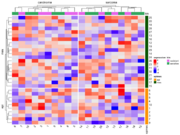 |
43 KB | Brb (talk | contribs) | <syntaxhighlight lang="rsplus"> # Simulate data library(ComplexHeatmap) ng <- 30; ns <- 20 set.seed(1) mat <- matrix(rnorm(ng*ns), nr=ng, nc=ns) colnames(mat) <- 1:ns rownames(mat) <- 1:ng # color bar on RHS ind_e <- 1:round(ng/3) ind_m <- (1+round(ng/3)):ng epimes <- rep(c("epi", "mes"), c(length(ind_e), length(ind_m))) row_ha <- rowAnnotation(epimes = epimes, col = list(epimes = c("epi" = "orange", "mes" = "darkgreen"))) # color bar on Top tumortype <- rep(c("carcinoma", "sarcoma"... | 1 |
| 11:15, 8 January 2023 | Reorder.dendrogram.png (file) |  |
10 KB | Brb (talk | contribs) | <pre> set.seed(123) x <- rnorm(20) hc <- hclust(dist(x)) dd <- as.dendrogram(hc) par(mfrow=c(3, 1)) plot(dd, main = "random dendrogram 'dd'") # not the same as reorder(dd, 1:20) plot(reorder(dd, 20:1), main = 'reorder(dd, 20:1, sum)') plot(reorder(dd, 20:1, mean), main = 'reorder(dd, 20:1, mean)') </pre> | 1 |
| 11:01, 12 January 2023 | GseaTable.png (file) |  |
81 KB | Brb (talk | contribs) | <pre> data(examplePathways) data(exampleRanks) fgseaRes <- fgsea(examplePathways, exampleRanks, nperm=1000, minSize=15, maxSize=100) # I pick 5 pathways with + NES and 5 pathways with - NES. fgseaRes[order(pval), ][62:71, c('pathway', 'NES')] # pathway NES # 1: 5992282_ECM_proteoglycans 1.984081 # 2: 5992219_Regulation_of_cholesterol_biosynthesis_by_SREBP_SREBF_ 1.95... | 1 |
| 11:18, 12 January 2023 | GseaTable2.png (file) |  |
81 KB | Brb (talk | contribs) | An example of a plot from 10 non-enriched pathways. <pre> data(examplePathways) data(exampleRanks) fgseaRes <- fgsea(examplePathways, exampleRanks, nperm=1000, minSize=15, maxSize=100) fgseaRes[order(pval, decreasing = T),][1:10, c('NES', 'pval')] # NES pval # 1: -0.4050950 1.0000000 # 2: -0.4050950 1.0000000 # 3: -0.4966664 0.9932584 # 4: 0.4804114 0.9870610 # 5: 0.4804114 0.9870610 # 6: 0.4804114 0.9870610 # 7: 0.4804114 0.9870610 # 8: 0.4955139 0.9854... | 1 |
| 20:50, 14 January 2023 | RbdTreat.png (file) | 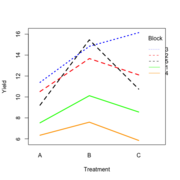 |
32 KB | Brb (talk | contribs) | <pre> set.seed(1234) block <- as.factor(rep(1:5, each=6)) treatment <- rep(c("A","B","C"),5) block_shift <- rnorm(5, mean = 0, sd = 2) treatment_shift <- c(A=0, B=4, C=2) random_effect <- rnorm(30, mean = 0, sd = 1) yield <- rnorm(30, mean = 10, sd = 2) + treatment_shift[as.integer(factor(treatment))] + block_shift[as.numeric(block)] + random_effect data <- data.frame(block, treatment, yield) summary(fm1 <- aov(yield ~ treatment + block, data = data)) # Df Sum Sq Mean Sq... | 1 |
| 20:51, 14 January 2023 | RbdBlock.png (file) | 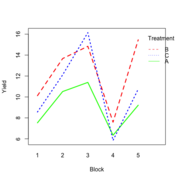 |
34 KB | Brb (talk | contribs) | <pre> set.seed(1234) block <- as.factor(rep(1:5, each=6)) treatment <- rep(c("A","B","C"),5) block_shift <- rnorm(5, mean = 0, sd = 2) treatment_shift <- c(A=0, B=4, C=2) random_effect <- rnorm(30, mean = 0, sd = 1) yield <- rnorm(30, mean = 10, sd = 2) + treatment_shift[as.integer(factor(treatment))] + block_shift[as.numeric(block)] + random_effect data <- data.frame(block, treatment, yield) summary(fm1 <- aov(yield ~ treatment + block, data = data)) # Df Sum Sq Mean Sq... | 1 |
| 21:26, 14 January 2023 | RbdGeom.png (file) | 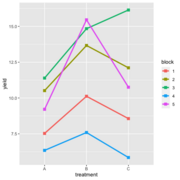 |
41 KB | Brb (talk | contribs) | <pre> require(ggplot2) aggregate( .~ treatment +block,FUN=median, data = data) |> ggplot(aes(treatment, yield)) + geom_line(aes(group = block, color = block), linewidth = 1.2) + geom_point(aes(color = block), shape = 15, size=2.6) </pre> | 1 |
| 09:21, 25 January 2023 | VscodeEnergy.png (file) |  |
116 KB | Brb (talk | contribs) | 1 | |
| 19:34, 28 January 2023 | Pca directly.png (file) | 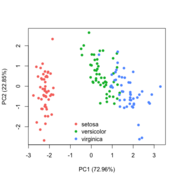 |
40 KB | Brb (talk | contribs) | 1 | |
| 19:34, 28 January 2023 | Pca autoplot.png (file) | 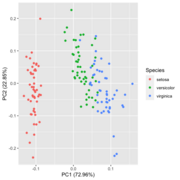 |
41 KB | Brb (talk | contribs) | 1 | |
| 20:01, 28 January 2023 | Pca factoextra.png (file) |  |
60 KB | Brb (talk | contribs) | 1 | |
| 10:08, 11 February 2023 | ClassPrediction.PNG (file) | 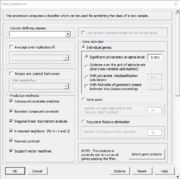 |
34 KB | Brb (talk | contribs) | 1 | |
| 10:10, 11 February 2023 | ClassPredictionOptions.PNG (file) | 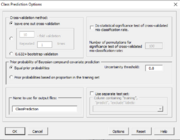 |
21 KB | Brb (talk | contribs) | 1 | |
| 10:11, 11 February 2023 | MultipleProbes.PNG (file) | 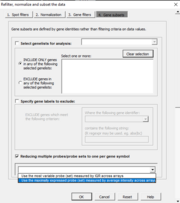 |
28 KB | Brb (talk | contribs) | 1 | |
| 15:01, 8 March 2023 | Pca directly2.png (file) | 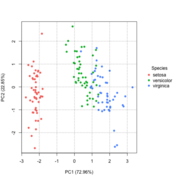 |
35 KB | Brb (talk | contribs) | 1 | |
| 09:44, 11 March 2023 | RStudioVisualMode.png (file) |  |
10 KB | Brb (talk | contribs) | 1 | |
| 15:58, 17 March 2023 | Svg4.svg (file) | 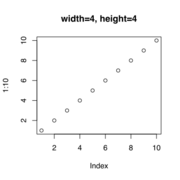 |
33 KB | Brb (talk | contribs) | <pre> svg("svg4.svg", width=4, height=4) plot(1:10, main="width=4, height=4") dev.off() </pre> | 1 |
| 16:47, 23 April 2023 | Sleepstudy.png (file) | 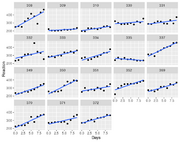 |
59 KB | Brb (talk | contribs) | <syntaxhighlight lang='rsplus'> sleepstudy %>% ggplot(aes(x=Days, y = Reaction)) + geom_point() + geom_smooth(method = "lm", se = FALSE) + facet_wrap(~Subject) </syntaxhighlight> | 1 |
| 20:07, 25 April 2023 | Losslesscut.png (file) |  |
323 KB | Brb (talk | contribs) | 1 | |
| 10:40, 9 May 2023 | Cbioportal cptac.png (file) |  |
120 KB | Brb (talk | contribs) | 1 | |
| 12:52, 9 May 2023 | Ggplotbarplot.png (file) |  |
23 KB | Brb (talk | contribs) | 1 | |
| 10:08, 10 May 2023 | Rpalette.png (file) | 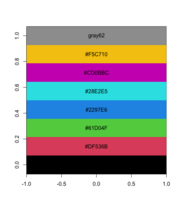 |
28 KB | Brb (talk | contribs) | <syntaxhighlight lang='rsplus'> pal <- palette() # [1] "black" "#DF536B" "#61D04F" "#2297E6" "#28E2E5" "#CD0BBC" "#F5C710" # [8] "gray62" pal_matrix <- matrix(seq_along(pal), nr=1) image(pal_matrix, col = pal, axes = FALSE) # 8 rows, 1 column, but no labeling # Starting from bottom, left. par()$usr # change with the data dim text(0, (par()$usr[4]-par()$usr[3])/8*c(0:7), labels = pal) </syntaxhighlight> | 1 |