Heatmap: Difference between revisions
| Line 1: | Line 1: | ||
= Clustering = | = Clustering = | ||
== Task View == | |||
https://cran.r-project.org/web/views/Cluster.html | |||
== k-means clustering == | == k-means clustering == | ||
* [http://varianceexplained.org/r/kmeans-free-lunch/ Assumptions], a post from varianceexplained.org. | * [http://varianceexplained.org/r/kmeans-free-lunch/ Assumptions], a post from varianceexplained.org. | ||
Revision as of 13:42, 30 October 2020
Clustering
Task View
https://cran.r-project.org/web/views/Cluster.html
k-means clustering
- Assumptions, a post from varianceexplained.org.
- UTILIZING K-MEANS TO EXTRACT COLOURS FROM YOUR FAVOURITE IMAGES
k-medoids/Partitioning Around Medoids (PAM)
- https://en.wikipedia.org/wiki/K-medoids
- https://www.rdocumentation.org/packages/cluster/versions/2.0.7-1/topics/pam
Number of clusters: Intraclass Correlation/ Intra Cluster Correlation (ICC)
- https://en.wikipedia.org/wiki/Intraclass_correlation
- ICC was used by Integrating gene expression and GO classification for PCA by preclustering by Hann et al 2010
Hierarchical clustering
For the kth cluster, define the Error Sum of Squares as [math]\displaystyle{ ESS_m = }[/math] sum of squared deviations (squared Euclidean distance) from the cluster centroid. [math]\displaystyle{ ESS_m = \sum_{l=1}^{n_m}\sum_{k=1}^p (x_{ml,k} - \bar{x}_{m,k})^2 }[/math] in which [math]\displaystyle{ \bar{x}_{m,k} = (1/n_m) \sum_{l=1}^{n_m} x_{ml,k} }[/math] the mean of the mth cluster for the k-th variable, [math]\displaystyle{ x_{ml,k} }[/math] being the score on the kth variable [math]\displaystyle{ (k=1,\dots,p) }[/math] for the l-th object [math]\displaystyle{ (l=1,\dots,n_m) }[/math] in the mth cluster [math]\displaystyle{ (m=1,\dots,g) }[/math].
If there are C clusters, define the Total Error Sum of Squares as Sum of Squares as [math]\displaystyle{ ESS = \sum_m ESS_m, m=1,\dots,C }[/math]
Consider the union of every possible pair of clusters.
Combine the 2 clusters whose combination combination results in the smallest increase in ESS.
Comments:
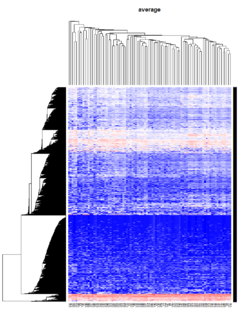
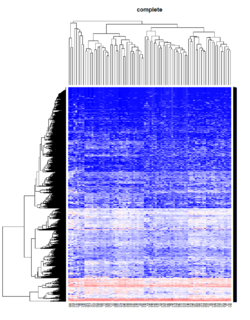
- The default linkage is "complete" in R.
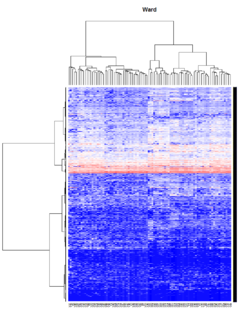
- Ward's method tends to join clusters with a small number of observations, and it is strongly biased toward producing clusters with the same shape and with roughly the same number of observations.
- It is also very sensitive to outliers. See Milligan (1980).
Take pomeroy data (7129 x 90) for an example:
library(gplots)
lr = read.table("C:/ArrayTools/Sample datasets/Pomeroy/Pomeroy -Project/NORMALIZEDLOGINTENSITY.txt")
lr = as.matrix(lr)
method = "average" # method <- "complete"; method <- "ward.D"; method <- "ward.D2"
hclust1 <- function(x) hclust(x, method= method)
heatmap.2(lr, col=bluered(75), hclustfun = hclust1, distfun = dist,
density.info="density", scale = "none",
key=FALSE, symkey=FALSE, trace="none",
main = method)
Density based clustering
http://www.r-exercises.com/2017/06/10/density-based-clustering-exercises/
Optimal number of clusters
- https://datascienceplus.com/finding-optimal-number-of-clusters/
- 10 Tips for Choosing the Optimal Number of Clusters
Benchmark clustering algorithms
Using clusterlab to benchmark clustering algorithms
You probably don't understand heatmaps
- http://www.opiniomics.org/you-probably-dont-understand-heatmaps/
- Effect of number of genes
Evaluate the effect of centering & scaling
1-correlation distance
Effect of centering and scaling on clustering of genes and samples in terms of distance. 'Yes' means the distance was changed compared to the baseline where no centering or scaling was applied.
| clustering genes | clustering samples | |
|---|---|---|
| centering on each genes | No | Yes* |
| scaling on each genes | No | Yes* |
- Note: Cor(X, Y) = Cor(X + constant scalar, Y). If the constant is not a scalar, the equation won't hold. Or think about plotting data in a 2 dimension space. If the X data has a constant shift for all observations/genes, then the linear correlation won't be changed.
Euclidean distance
| clustering genes | clustering samples | |
|---|---|---|
| centering on each genes | Yes | No1 |
| scaling on each genes | Yes | Yes2 |
Note
- [math]\displaystyle{ \sum(X_i - Y_i)^2 = \sum(X_i-c_i - (Y_i-c_i))^2 }[/math]
- [math]\displaystyle{ \sum(X_i - Y_i)^2 \neq \sum(X_i/c_i - Y_i/c_i)^2 }[/math]
Supporting R code
1. 1-Corr distance
source("http://www.bioconductor.org/biocLite.R")
biocLite("limma"); biocLite("ALL")
library(limma); library(ALL)
data(ALL)
eset <- ALL[, ALL$mol.biol %in% c("BCR/ABL", "ALL1/AF4")]
f <- factor(as.character(eset$mol.biol))
design <- model.matrix(~f)
fit <- eBayes(lmFit(eset,design))
selected <- p.adjust(fit$p.value[, 2]) < 0.05
esetSel <- eset [selected, ] # 165 x 47
heatmap(exprs(esetSel))
esetSel2 <- esetSel[sample(1:nrow(esetSel), 20), sample(1:ncol(esetSel), 10)] # 20 x 10
dist.no <- 1-cor(t(as.matrix(esetSel2)))
dist.mean <- 1-cor(t(sweep(as.matrix(esetSel2), 1L, rowMeans(as.matrix(esetSel2))))) # gene variance has not changed!
dist.median <- 1-cor(t(sweep(as.matrix(esetSel2), 1L, apply(esetSel2, 1, median))))
range(dist.no - dist.mean) # [1] -1.110223e-16 0.000000e+00
range(dist.no - dist.median) # [1] -1.110223e-16 0.000000e+00
range(dist.mean - dist.median) # [1] 0 0
So centering (no matter which measure: mean, median, ...) genes won't affect 1-corr distance of genes.
dist.mean <- 1-cor(t(sweep(as.matrix(esetSel2), 1L, rowMeans(as.matrix(esetSel2)), "/"))) dist.median <- 1-cor(t(sweep(as.matrix(esetSel2), 1L, apply(esetSel2, 1, median), "/"))) range(dist.no - dist.mean) # [1] -8.881784e-16 6.661338e-16 range(dist.no - dist.median) # [1] -6.661338e-16 6.661338e-16 range(dist.mean - dist.median) # [1] -1.110223e-15 1.554312e-15
So scaling after centering (no matter what measures: mean, median,...) won't affect 1-corr distance of genes.
How about centering / scaling genes on array clustering?
dist.no <- 1-cor(as.matrix(esetSel2)) dist.mean <- 1-cor(sweep(as.matrix(esetSel2), 1L, rowMeans(as.matrix(esetSel2)))) # array variance has changed! dist.median <- 1-cor(sweep(as.matrix(esetSel2), 1L, apply(esetSel2, 1, median))) range(dist.no - dist.mean) # [1] -1.547086 0.000000 range(dist.no - dist.median) # [1] -1.483427 0.000000 range(dist.mean - dist.median) # [1] -0.5283601 0.6164602 dist.mean <- 1-cor(sweep(as.matrix(esetSel2), 1L, rowMeans(as.matrix(esetSel2)), "/")) dist.median <- 1-cor(sweep(as.matrix(esetSel2), 1L, apply(esetSel2, 1, median), "/")) range(dist.no - dist.mean) # [1] -1.477407 0.000000 range(dist.no - dist.median) # [1] -1.349419 0.000000 range(dist.mean - dist.median) # [1] -0.5419534 0.6269875
2. Euclidean distance
dist.no <- dist(as.matrix(esetSel2)) dist.mean <- dist(sweep(as.matrix(esetSel2), 1L, rowMeans(as.matrix(esetSel2)))) dist.median <- dist(sweep(as.matrix(esetSel2), 1L, apply(esetSel2, 1, median))) range(dist.no - dist.mean) # [1] 7.198864e-05 2.193487e+01 range(dist.no - dist.median) # [1] -0.3715231 21.9320846 range(dist.mean - dist.median) # [1] -0.923717629 -0.000088385
Centering does affect the Euclidean distance.
dist.mean <- dist(sweep(as.matrix(esetSel2), 1L, rowMeans(as.matrix(esetSel2)), "/")) dist.median <- dist(sweep(as.matrix(esetSel2), 1L, apply(esetSel2, 1, median), "/")) range(dist.no - dist.mean) # [1] 0.7005071 24.0698991 range(dist.no - dist.median) # [1] 0.636749 24.068920 range(dist.mean - dist.median) # [1] -0.22122869 0.02906131
And scaling affects Euclidean distance too.
How about centering / scaling genes on array clustering?
dist.no <- dist(t(as.matrix(esetSel2))) dist.mean <- dist(t(sweep(as.matrix(esetSel2), 1L, rowMeans(as.matrix(esetSel2))))) dist.median <- dist(t(sweep(as.matrix(esetSel2), 1L, apply(esetSel2, 1, median)))) range(dist.no - dist.mean) # 0 0 range(dist.no - dist.median) # 0 0 range(dist.mean - dist.median) # 0 0 dist.mean <- dist(t(sweep(as.matrix(esetSel2), 1L, rowMeans(as.matrix(esetSel2)), "/"))) dist.median <- dist(t(sweep(as.matrix(esetSel2), 1L, apply(esetSel2, 1, median), "/"))) range(dist.no - dist.mean) # [1] 1.698960 9.383789 range(dist.no - dist.median) # [1] 1.683028 9.311603 range(dist.mean - dist.median) # [1] -0.09139173 0.02546394
Euclidean distance and Pearson correlation relationship
http://analytictech.com/mb876/handouts/distance_and_correlation.htm. In summary,
[math]\displaystyle{ r(X, Y) = 1 - \frac{d^2(X, Y)}{2n} }[/math]
where [math]\displaystyle{ r(X, Y) }[/math] is the Pearson correlation of variables X and Y and [math]\displaystyle{ d^2(X, Y) }[/math] is the squared Euclidean distance of X and Y.
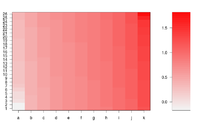
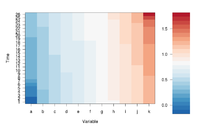
Simple image plot using image() function
https://chitchatr.wordpress.com/2010/07/01/matrix-plots-in-r-a-neat-way-to-display-three-variables/
### Create Matrix plot using colors to fill grid
# Create matrix. Using random values for this example.
rand <- rnorm(286, 0.8, 0.3)
mat <- matrix(sort(rand), nr=26)
dim(mat) # Check dimensions
# Create x and y labels
yLabels <- seq(1, 26, 1)
xLabels <- c("a", "b", "c", "d", "e", "f", "g", "h", "i",
"j", "k");
# Set min and max values of rand
min <- min(rand, na.rm=T)
max <- max(rand, na.rm=T)
# Red and green range from 0 to 1 while Blue ranges from 1 to 0
ColorRamp <- rgb(seq(0.95,0.99,length=50), # Red
seq(0.95,0.05,length=50), # Green
seq(0.95,0.05,length=50)) # Blue
ColorLevels <- seq(min, max, length=length(ColorRamp))
# Set layout. We are going to include a colorbar next to plot.
layout(matrix(data=c(1,2), nrow=1, ncol=2), widths=c(4,1),
heights=c(1,1))
#plotting margins. These seem to work well for me.
par(mar = c(5,5,2.5,1), font = 2)
# Plot it up!
image(1:ncol(mat), 1:nrow(mat), t(mat),
col=ColorRamp, xlab="Variable", ylab="Time",
axes=FALSE, zlim=c(min,max),
main= NA)
# Now annotate the plot
box()
axis(side = 1, at=seq(1,length(xLabels),1), labels=xLabels,
cex.axis=1.0)
axis(side = 2, at=seq(1,length(yLabels),1), labels=yLabels, las= 1,
cex.axis=1)
# Add colorbar to second plot region
par(mar = c(3,2.5,2.5,2))
image(1, ColorLevels,
matrix(data=ColorLevels, ncol=length(ColorLevels),nrow=1),
col=ColorRamp,xlab="",ylab="",xaxt="n", las = 1)
If we define ColorRamp variable using the following way, we will get the 2nd plot.
require(RColorBrewer) # get brewer.pal() ColorRamp <- colorRampPalette( rev(brewer.pal(9, "RdBu")) )(25)
Note that
- colorRampPalette() is an R's built-in function. It interpolate a set of given colors to create new color palettes. The return is a function that takes an integer argument (the required number of colors) and returns a character vector of colors (see rgb()) interpolating the given sequence (similar to heat.colors() or terrain.colors()).
# An example of showing 50 shades of grey in R
greys <- grep("^grey", colours(), value = TRUE)
length(greys)
# [1] 102
shadesOfGrey <- colorRampPalette(c("grey0", "grey100"))
shadesOfGrey(2)
# [1] "#000000" "#FFFFFF"
# And 50 shades of grey?
fiftyGreys <- shadesOfGrey(50)
mat <- matrix(rep(1:50, each = 50))
image(mat, axes = FALSE, col = fiftyGreys)
box()
- (For dual channel data) brewer.pal(9, "RdBu") creates a diverging palette based on "RdBu" with 9 colors. See help(brewer.pal, package="RColorBrewer") for a list of palette name. The meaning of the palette name can be found on colorbrew2.org website.
- (For single channel data) brewer.pal(9, "Blues") is good. See an example.
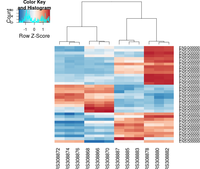
gplots package
The following example is extracted from DESeq2 package.
## ----loadDESeq2, echo=FALSE----------------------------------------------
# in order to print version number below
library("DESeq2")
## ----loadExonsByGene, echo=FALSE-----------------------------------------
library("parathyroidSE")
library("GenomicFeatures")
data(exonsByGene)
## ----locateFiles, echo=FALSE---------------------------------------------
bamDir <- system.file("extdata",package="parathyroidSE",mustWork=TRUE)
fls <- list.files(bamDir, pattern="bam$",full=TRUE)
## ----bamfilepaired-------------------------------------------------------
library( "Rsamtools" )
bamLst <- BamFileList( fls, yieldSize=100000 )
## ----sumOver-------------------------------------------------------------
library( "GenomicAlignments" )
se <- summarizeOverlaps( exonsByGene, bamLst,
mode="Union",
singleEnd=FALSE,
ignore.strand=TRUE,
fragments=TRUE )
## ----libraries-----------------------------------------------------------
library( "DESeq2" )
library( "parathyroidSE" )
## ----loadEcs-------------------------------------------------------------
data( "parathyroidGenesSE" )
se <- parathyroidGenesSE
colnames(se) <- se$run
## ----fromSE--------------------------------------------------------------
ddsFull <- DESeqDataSet( se, design = ~ patient + treatment )
## ----collapse------------------------------------------------------------
ddsCollapsed <- collapseReplicates( ddsFull,
groupby = ddsFull$sample,
run = ddsFull$run )
## ----subsetCols----------------------------------------------------------
dds <- ddsCollapsed[ , ddsCollapsed$time == "48h" ]
## ----subsetRows, echo=FALSE----------------------------------------------
idx <- which(rowSums(counts(dds)) > 0)[1:4000]
dds <- dds[idx,]
## ----runDESeq, cache=TRUE------------------------------------------------
dds <- DESeq(dds)
rld <- rlog( dds)
library( "genefilter" )
topVarGenes <- head( order( rowVars( assay(rld) ), decreasing=TRUE ), 35 )
## ----beginner_geneHeatmap, fig.width=9, fig.height=9---------------------
library(RColorBrewer)
library(gplots)
heatmap.2( assay(rld)[ topVarGenes, ], scale="row",
trace="none", dendrogram="column",
col = colorRampPalette( rev(brewer.pal(9, "RdBu")) )(255))
Self-defined distance/linkage method
heatmap.2(..., hclustfun = function(x) hclust(x,method = 'ward.D2'), ...)
Change breaks in scale
https://stackoverflow.com/questions/17820143/how-to-change-heatmap-2-color-range-in-r
Con: it'll be difficult to interpret the heatmap
Color labels
https://stackoverflow.com/questions/13206335/color-labels-text-in-r-heatmap
Moving colorkey
White strips (artifacts)
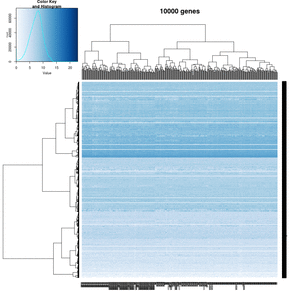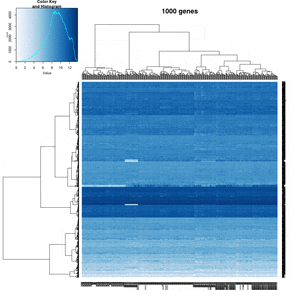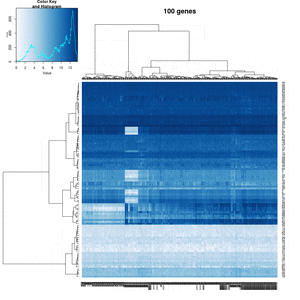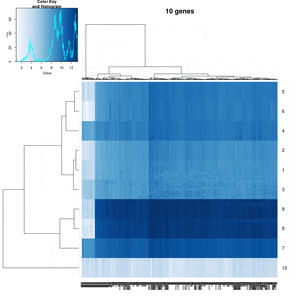
On my Linux (Dell Precision T3500, NVIDIA GF108GL, Quadro 600, 1920x1080), the heatmap shows several white strips when I set a resolution 800x800 (see the plot of 10000 genes shown below). Note if I set a higher resolution 1920x1920, the problem is gone but the color key map will be quite large and the text font will be small.
On MacBook Pro (integrated Intel Iris Pro, 2880x1800), there is no artifact even with 800x800 resolution.
How about saving the plots using
- a different format (eg tiff) or even the lossless compression option - not help
- Cairo package - works. Note that the default background is transparent.
Output from heatmap.2 examples
ggplot2 package
- https://learnr.wordpress.com/2010/01/26/ggplot2-quick-heatmap-plotting/
- http://www.kim-herzig.de/2014/06/19/heat-maps-with-ggplot2/
- http://is-r.tumblr.com/post/32387034930/simplest-possible-heatmap-with-ggplot2
- http://smartdatawithr.com/en/creating-heatmaps/#more-875 data values are shown in cells!
NMF package
aheatmap() function.
- http://nmf.r-forge.r-project.org/aheatmap.html
- http://www.gettinggeneticsdone.com/2015/04/r-user-group-recap-heatmaps-and-using.html
ComplexHeatmap
- Groups of heatmaps
- The color argument can contain a mapping function or a vector of colors. The circlize package (from the same package author) can be used.
- A simple tutorial for a complex ComplexHeatmap
Pros
- Annotation of classes for a new variable.
Correlation matrix
The following code will guarantee the heatmap has a diagonal in the direction of top-left and bottom-right. The row dendrogram will be flipped automatically. There is no need to use cluster_rows = rev(mydend).
mcor <- cor(t(lograt))
colnames(mcor) <- rownames(mcor) <- 1:ncol(mcor)
mydend <- as.dendrogram(hclust(as.dist(1-mcor)))
Heatmap( mcor, cluster_columns = mydend, cluster_rows = mydend,
row_dend_reorder = FALSE, column_dend_reorder = FALSE,
row_names_gp = gpar(fontsize = 6),
column_names_gp = gpar(fontsize = 6),
column_title = "", name = "value")
OncoPrint
- https://jokergoo.github.io/ComplexHeatmap-reference/book/oncoprint.html
- http://qiubio.com/new/book/chapter-08/#oncoprint
- http://blog.thehyve.nl/blog/downloading-data-from-the-cbioportal-oncoprint-view
tidyHeatmap
https://github.com/stemangiola/tidyHeatmap
Correlation heatmap
Customizable correlation heatmaps in R using purrr and ggplot2
ggcorrplot
https://cran.r-project.org/web/packages/ggcorrplot/index.html
SubtypeDrug: Prioritization of Candidate Cancer Subtype Specific Drugs
https://cran.r-project.org/web/packages/SubtypeDrug/index.html
pheatmap
- http://wiki.bits.vib.be/index.php/Use_pheatmap_to_draw_heat_maps_in_R
- Making a heatmap in R with the pheatmap package
- https://stackoverflow.com/questions/13087555/heatmap-in-r-how-to-resize-columns-labels
- The package was used on the book Modern Statistics for Modern Biology
- Examples: PDAC data
What is special?
- the color scheme is grey-blue to orange-red. pheatmap() default options, colorRampPalette in R
- able to include class labels in samples and/or genes
- the color key is thin and placed on the RHS (prettier for publications though it could miss information)
- borders for each cell are included (not necessary)
fheatmap
(archived)
corrplot
This package is used for visualization of correlation matrix. See its vignette and Visualize correlation matrix using correlogram.
Heatmap in the terminal
Interactive heatmaps
d3heatmap
A package generates interactive heatmaps using d3.js and htmlwidgets.
The package let you
- Highlight rows/columns by clicking axis labels
- Click and drag over colormap to zoom in (click on colormap to zoom out)
- Optional clustering and dendrograms, courtesy of base::heatmap
The following screenshots shows 3 features.
- Shows the row/column/value under the mouse cursor
- Zoom in a region (click on the zoom-in image will bring back the original heatmap)
- Highlight a row or a column (click the label of another row will highlight another row. Click the same label again will bring back the original image)
heatmaply
- http://moderndata.plot.ly/interactive-heat-maps-for-r/
- heatmaply: an R package for creating interactive cluster heatmaps for online publishing Bioinformatics 2017.
This package extends the plotly engine to heatmaps, allowing you to inspect certain values of the data matrix by hovering the mouse over a cell. You can also zoom into a region of the heatmap by drawing a rectangle over an area of your choice.
Installing this package requires to compile some dependent package.
The return object is heatmaply is 'plotly' and 'htmlwidget'. It does not return the ordering of rows/columns. It can not control whether to do clustering (d3heatmap package is better at this).
canvasXpress
- https://cran.r-project.org/web/packages/canvasXpress/index.html
- https://canvasxpress.org/html/heatmap-9.html
Dendrogram
Flip dendrogram
https://www.biostars.org/p/279775/
Color labels
library(RColorBrewer)
# matrix contains genomics-style data where columns are samples
# (if otherwise remove the transposition below)
# labels is a factor variable going along the columns of matrix
plotHclustColors <- function(matrix,labels,...) {
colnames(matrix) <- labels
d <- dist(t(matrix))
hc <- hclust(d, method = "ward.D2")
labelColors <- brewer.pal(nlevels(labels),"Set1")
colLab <- function(n) {
if (is.leaf(n)) {
a <- attributes(n)
labCol <- labelColors[which(levels(labels) == a$label)]
attr(n, "nodePar") <- c(a$nodePar, lab.col=labCol)
}
n
}
clusDendro <- dendrapply(as.dendrogram(hc), colLab)
# I change cex because there are lots of samples
op <- par(mar=c(5,3,1,.5)+.1, cex=.3)
plot(clusDendro,...)
par(op)
}
plotHclustColors(genedata, pheno)
Dendrogram with covariates
https://web.stanford.edu/~hastie/TALKS/barossa.pdf#page=41
dendextend package
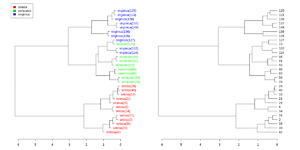
The package has a function tanglegram() to compare two trees of hierarchical clusterings. See this post and its vignette.
Simplified from dendextend's vignette.
library(dendextend)
set.seed(1234)
iris <- datasets::iris[sample(150, 30), ] # subset for better view
iris2 <- iris[, -5] # data
species_labels <- iris[, 5] # group for coloring
hc_iris <- hclust(dist(iris2), method = "complete")
iris_species <- levels(species_labels)
dend <- as.dendrogram(hc_iris)
colorCodes <- c("red", "green", "blue")
labels_colors(dend) <- colorCodes[as.numeric(species_labels)][order.dendrogram(dend)]
labels(dend) <- paste0(as.character(species_labels)[order.dendrogram(dend)],
"(", labels(dend), ")")
# We hang the dendrogram a bit:
dend <- hang.dendrogram(dend, hang_height=0.1)
dend <- set(dend, "labels_cex", 1.0)
png("~/Downloads/iris_dextend.png", width = 1200, height = 600)
par(mfrow=c(1,2), mar = c(3,3,1,7))
plot(dend, main = "", horiz = TRUE)
legend("topleft", legend = iris_species, fill = colorCodes)
par(mar=c(3,1,1,5))
plot(as.dendrogram(hc_iris),horiz=TRUE)
dev.off()
Colors
A quick introduction to using color in density plots
http://sharpsightlabs.com/blog/quick-intro-color-density-plot/
display.brewer.pal() and brewer.pal() functions
While brewer.pal() will return colors (in hex) for a certain palette, display.brew.pal() can display the colors on a graphical device.
library(RColorBrewer) display.brewer.pal(11, "BrBG") # Alternative colors used in correlation matrix display.brewer.pal(9, "Set1") # Up to 9 classes are allowed
Missing data
In the dynamic heatmap tool of BRB-ArrayTools, the missing data is represented by the gray color.
Papers
Healthcare Access and Quality Index - Lancet
http://www.thelancet.com/pdfs/journals/lancet/PIIS0140-6736(17)30818-8.pdf