GSEA: Difference between revisions
| Line 708: | Line 708: | ||
* [http://bioconductor.org/packages/release/bioc/html/phantasus.html phantasus] Visual and interactive gene expression analysis. read.gct(), [https://rdrr.io/bioc/phantasus/man/write.gct.html write.gct()], getGSE(), loadGEO(), gseaPlot(). | * [http://bioconductor.org/packages/release/bioc/html/phantasus.html phantasus] Visual and interactive gene expression analysis. read.gct(), [https://rdrr.io/bioc/phantasus/man/write.gct.html write.gct()], getGSE(), loadGEO(), gseaPlot(). | ||
* [https://artyomovlab.wustl.edu/phantasus/phantasus-tutorial.html Using Phantasus application] | * [https://artyomovlab.wustl.edu/phantasus/phantasus-tutorial.html Using Phantasus application] | ||
= PhenoExam = | |||
[https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-022-05122-x PhenoExam: gene set analyses through integration of different phenotype databa] | |||
= Selected papers = | = Selected papers = | ||
Revision as of 15:28, 2 January 2023
Basic
https://en.wikipedia.org/wiki/Gene_set_enrichment_analysis
Determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states
- https://www.bioconductor.org/help/course-materials/2015/SeattleApr2015/E_GeneSetEnrichment.html, https://www.bioconductor.org/help/course-materials/2009/SeattleApr09/gsea/HyperG_Lecture.pdf. For hypergeometric test, it is
- finding Biocarta or KEGG pathways significantly enriched in the user's gene list
- Are DE genes in the set more common than DE genes not in the set?.
- Are selected genes more often in the GO category than expected by chance? (one-tailed)
We can draw a 2x2 table or a venn diagram to see the diagram.
In gene set Yes No DE Yes No
- Gene List Enrichment Analysis dhyper(), binom.test(), fisher.test().
- Pathway Commons
- Gene set analysis methods: statistical models and methodological differences 2014
- http://software.broadinstitute.org/gsea/index.jsp, Subramanian, et al 2005 paper
- HOW TO PERFORM GSEA - A tutorial on gene set enrichment analysis for RNA-seq (video)
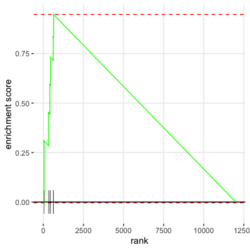
- Algorithm. GSEA walks down the ranked list of genes, increasing a running-sum statistic when a gene belongs to the set and decreasing it when the gene does not.
- Interpretation of 3 enrichment plots. The 1st plot (ES on y-axis) tells you how over or under expressed is your gene respect to the ranked list. The 2nd part of the graph (barcode-like) shows where the members of the gene set appear in the ranked list of genes. The 3rd graph (y=ranked list metric, x=rank) shows how your metric is distributed along the list.
- slides with formulas.
- What does a negative enrichment score mean? A negative NES will indicate that the genes in the set S will be mostly at the bottom of your list L.
- GSEA R Implementation from GSEA-MSigDB.
- READMe.
- Using GSEA.1.0.R
- Statistical power of gene-set enrichment analysis is a function of gene set correlation structure by SWANSON 2017
- Towards a gold standard for benchmarking gene set enrichment analysis, GSEABenchmarkeR package
- clusterProfiler package and the online book.
- Gene-set Enrichment with Regularized Regression Fang 2019
- msigdbr package. MSigDB Gene Sets for Multiple Organisms in a Tidy Data Format.
- Best method/package for Gene Set Enrichment Analysis in R? and the gage package
- ES could be negative; see Genome 559: Introduction to Statistical and Computational Genomics
- multiGSEA: a GSEA-based pathway enrichment analysis for multi-omics data
- How to use DAVID for functional annotation of genes, Using DAVID for Functional Enrichment Analysis in a Set of Genes (Part 1), (Part 2) (video)
Two categories of GSEA procedures:
- Competitive: compare genes in the test set relative to all other genes.
- Self-contained: whether the gene-set is more DE than one were to expect under the null of no association between two phenotype conditions (without reference to other genes in the genome). For example the method by Jiang & Gentleman Bioinformatics 2007
See also BRB-ArrayTools -> GSEA.
Interpretation
- Gene Set Enrichment Analysis (GSEA) takes an alternative approach : it focuses on cumulative changes in expression of multiple genes as a group (belonging to a same gene-set/pahtway), which shifts the focus from individual genes to groups of genes. See this.
- XXX class is associated with the XXX gene sets (GSVA vignette)
- XXX subtype is characterized by the expression of XXX markers, thus we expect it to correlate with the XXX gene set (GSVA vignette)
- The XXX subtype shows high expression of XXX genes, thus the XXX gene sets is highly enriched for this group (GSVA vignette)
- Negative ES Interpretation.
- A positive enrichment score should always reflect enrichment on the positive side of the zero cross (although not necessarily all genes on the positive side) and be enrichment in whichever phenotype was selected to be first in the comparison in the Phenotype selection dialogue. And vice versa, with a negative enrichment score reflecting enrichment in the genes on the negative side of the ranked list.
- GSEA is known to have some issues with highly skewed gene distributions, but that shouldn't affect the raw enrichment scores, just NES and significance calculations when GSEA runs in to a condition where its only sampled from one side of the distribution.
- Enrichment Score Interpretation
- If, for example, you provide a gene list ranked by a combination of fold change and p-value (e.g., sign(FC) * log10(pvalue)), then the positive scores are associated with upregulated genes and negative scores are associated with downregulated genes.
- fgsea paper
- The more positive is the value of sr(p) the more enriched the gene set is in the positively-regulated genes (with Si > 0). Accordingly, negative sr(p) corresponds to enrichment in the negatively regulated genes.
Calculation
- Subramanian paper
- https://youtu.be/bT00oJh2x_4
- *pathwaycommons.org
- Compute cumulative sum over ranked genes. MD Anderson lecture
- Increase sum when gene in set, decrease it otherwise. That is, +1/-1 weights in cumulative sum is used to represent whether genes are in the interested gene set.
- Magnitude of increment depends on correlation of gene with phenotype.
- Record the maximum deviation from zero as the enrichment score
FDR cutoff
Why does GSEA use a false discovery rate (FDR) of 0.25 rather than the more classic 0.05?
piano
- piano package. Platform for integrative analysis of omics data.
- Available GSA methods.
- The main function is runGSA(). It contains a wide range of GSEA methods like maxmean, fisher, gsea, fgsea, etc.
- Conceptual overview of the runGSA workflow
- It was used in Xeva paper.
edgeR
Over-representation analysis. ?goana and ?kegga. See UserGuide 2.14 Gene ontology (GO) and pathway analysis.
fgsea
fgsea package and download stat
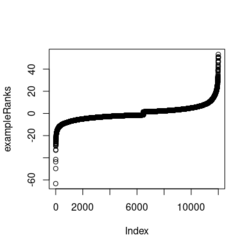
- ?exampleRanks gives a hint about how it was obtained. It is actually the t-statistic, not logFC.
... fit2 <- eBayes(fit2) de <- data.table(topTable(fit2, adjust.method="BH", number=12000, sort.by = "B"), keep.rownames = TRUE) colnames(de) plot(de$t, exampleRanks[match(de$rn, names(exampleRanks))]) # 45 degrees straight line range(de$t) # [1] -62.22877 52.59930 range(exampleRanks) # [1] -63.33703 53.28400 range(de$logFC) # [1] -11.56067 12.57562 - Paper is still on biorxiv only. Scholar.google.com. Computer tech lab in Russia.
- Documentation
- ?collapsePathways. Collapse list of enriched pathways to independent ones.
- DESeq results to pathways in 60 Seconds with the fgsea package
- Performing GSEA using MSigDB gene sets in R
- Using the fast preranked gene set enrichment analysis (fgsea) package
Are fgsea and Broad Institute GSEA equivalent
Are fgsea and Broad Institute GSEA equivalent?
Examples
- vignette,
- plotEnrichment() for a single pathway (including the output plot). No need to run fgsea(). Source code. Enrichment score (ES) on the plot is calculated by calcGseaStat()$res. The ES value is the same as the one shown in plotGseaTable() though plotEnrichment does not return it.
- plotGseaTable() for a bunch of selected pathways (including the output plot). The range of NES is not [-1, 1] but ES seems to be in [-1, 1]. The example sort pathways by NES but it can be adapted to sort by pval. Source code. Also ES and NES are not in the same order (see below) though we cannot tell it from the small plot.
- Basic usage
library(fgsea) library(ggplot2) data(examplePathways) # a list of length 1457 (pathways) data(exampleRanks) # a vector of length 12000 (genes) set.seed(42) fgseaRes <- fgsea(pathways = examplePathways, stats = exampleRanks, minSize = 4, # default minSize=1, maxSize=Inf maxSize =10) # I used a very small maxSize in order to see details later # So the results here can't be compared with the default class(fgseaRes) # [1] "data.table" "data.frame" dim(fgseaRes) # [1] 386 8 fgseaRes[1:2, ] # pathway pval padj # 1: 1368092_Rora_activates_gene_expression 0.8630705 0.9386018 # 2: 1368110_Bmal1:Clock,Npas2_activates_circadian_gene_expression 0.4312268 0.8159487 # log2err ES NES size leadingEdge # 1: 0.05412006 -0.3087414 -0.6545067 5 11865,12753,328572,20787 # 2: 0.08312913 0.4209054 1.0360288 9 20893,59027,19883 sapply(examplePathways, length) |> range() # [1] 1 2366 # Question: minSize, maxSize mean 'matched' genes? Can't verify sum(sapply(examplePathways, length) < 3) # [1] 104 sum(sapply(examplePathways, length) > 10) # [1] 939 1457 - 104 - 939 # [1] 414 examplePathways2 <- lapply(examplePathways, function(x) x[x %in% names(exampleRanks)]) sapply(examplePathways2, length) |> range() # [1] 0 968 sum(sapply(examplePathways2, length) < 3) # [1] 232 sum(sapply(examplePathways2, length) > 10) # [1] 730 1457 - 232 - 730 # [1] 495 range(fgseaRes$ES) # [1] -0.8442416 0.9488163 range(fgseaRes$NES) # [1] -2.020695 2.075729 order(fgseaRes$ES)[1:5] # [1] 75 289 320 249 312 order(fgseaRes$NES)[1:5] # [1] 289 75 102 320 200 # choose the top gene set in order to zoom in & see the detail head(fgseaRes[order(pval), ], 1) # pathway pval padj log2err ES # 1: 5991601_Phosphorylation_of_Emi1 2.461461e-07 9.501241e-05 0.6749629 0.9472236 # NES size leadingEdge # 1: 2.082967 6 107995,12534,18817,67141,268697,56371 plotEnrichment(examplePathways[["5991601_Phosphorylation_of_Emi1"]], exampleRanks) # 12000 total genes though only 6 genes are matched in this top gene set. debug(plotEnrichment) # Browse[2]> toPlot # x y # 1 0 0.000000000 # 2 47 -0.003918626 # 3 48 0.308356695 # 4 322 0.285511939 # 5 323 0.452415897 # 6 407 0.445412396 # 7 408 0.593677191 # 8 447 0.590425565 # 9 448 0.730277162 # 10 617 0.716186784 # 11 618 0.833696389 # 12 638 0.832028888 # 13 639 0.947223612 # 14 12001 0.000000000 examplePathways[["5991601_Phosphorylation_of_Emi1"]] # [1] "12534" "18817" "56371" "67141" "107995" "268697" # [7] "434175" "102643276" match(examplePathways[["5991601_Phosphorylation_of_Emi1"]], names(exampleRanks)) # [1] 11678 11593 11362 11553 11953 11383 NA NA # Since there are 12000 total genes and exampleRanks have been sorted, # these matched genes have very high ranks. # See the plot below. length(exampleRanks) # [1] 12000 exampleRanks[1:10] # 170942 109711 18124 12775 72148 16010 11931 # -63.33703 -49.74779 -43.63878 -41.51889 -33.26039 -32.77626 -29.42328 # 13849 241230 665113 # -28.83475 -28.65111 -27.81583 sum(exampleRanks < 0) # [1] 6469 sum(exampleRanks > 0) # [1] 5531 plot(exampleRanks)There are 6*2 points (excluding the 0 value at the starting and end) in the figure.
- Understand plotEnrichment() function. Plot shape (concave or convex) requires the stats of genes from both in- and out- of the gene set. This affects the interpretation of the plot. The plot always starts with 0 and end with 0 in Y (enrichment score). plotEnrichment() does not return enrichment scores even it calculates them for the plot.
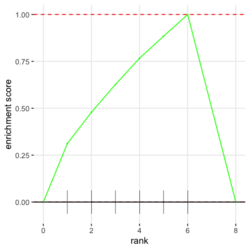
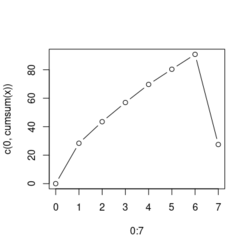
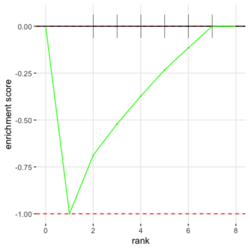
# Reduce the total number of genes i <- match(examplePathways[["5991601_Phosphorylation_of_Emi1"]], names(exampleRanks)) i <- na.omit(i) # Exclude 2 genes in the pathways but not in the data if (FALSE) { # Total genes = pathway genes will not work? plotEnrichment(examplePathways[["5991601_Phosphorylation_of_Emi1"]], exampleRanks[i]) # Error: GSEA statistic is not defined when all genes are selected } # PS values in 'exampleRanks' object are sorted from the smallest to the largest # Case 1: include 1 down-regulated gene for our total genes plotEnrichment(examplePathways[["5991601_Phosphorylation_of_Emi1"]], exampleRanks[c(1,i)]) # saved as fgseaPlotSmall.png exampleRanks[c(1,i)] # 170942 12534 18817 56371 67141 107995 268697 # -63.33703 15.16265 13.46935 10.46504 12.70504 28.36914 10.67534 # # Rank and add +1/-1, # 28.36 15.16 13.46 12.70 10.67 10.46 -63.33 # +1 +1 +1 +1 +1 +1 -1 debug(plotEnrichment) # Browse[2]> toPlot # x y # 1 0 0.0000000 # 2 0 0.0000000 # 3 1 0.3122753 # 4 1 0.3122753 # 5 2 0.4791793 # 6 2 0.4791793 # 7 3 0.6274441 # 8 3 0.6274441 # 9 4 0.7672957 # 10 4 0.7672957 # 11 5 0.8848053 # 12 5 0.8848053 # 13 6 1.0000000 # 14 8 0.0000000 x <-c(28.36, 15.16, 13.46, 12.70, 10.67, 10.46, -63.33) plot(0:7, c(0, cumsum(x)), type = "b") # Case 2: include another one up-regulated gene (so all are up-reg) for our total genes # However, the largest gene (added) is not in the gene set. # It will make the plot starting from a negative value (ignore 0 from the very left) plotEnrichment(examplePathways[["5991601_Phosphorylation_of_Emi1"]], exampleRanks[c(i, 12000)]) # saved as fgseaPlotSmall2.png exampleRanks[c(i, 12000)] # 12534 18817 56371 67141 107995 268697 80876 # 15.16265 13.46935 10.46504 12.70504 28.36914 10.67534 53.28400 # # Rank and add +1/-1 # 53.28 28.36 15.16 13.46 12.70 10.67 10.46 # -1 +1 +1 +1 +1 +1 +1For figure 1 (case 1), it has 6 points. Figure 2 is the one I generate using plot() and exampleRanks. For figure 3 (case 2), it has 7 points (excluding the 0 value at the starting and end). The curve goes down first because the gene has the largest rank (statistic) and is not in the pathway. So the x-axis location is determined by the rank/statistics and going up or down on the y-axis is determined by whether the gene is in the pathway or not.
- Shifting exampleRanks values will change the enrichment scores because the directions of genes' statistics change?
plotEnrichment(examplePathways[[1]], exampleRanks) + labs(title="Programmed Cell Death") # like sin() with one period range(exampleRanks) # [1] -63.33703 53.28400 plotEnrichment(examplePathways[[1]], exampleRanks+64) + labs(title="Programmed Cell Death") # like a time series plot plotEnrichment(examplePathways[[1]], exampleRanks-54) + labs(title="Programmed Cell Death") # same problem if we shift exampleRanks to all negativeincluding NE, NES, pval, etc. So it is not purely rank-based.
set.seed(42) fgseaRes2 <- fgsea(pathways = examplePathways, stats = exampleRanks+2, # OR 2*(exampleRanks+2) minSize = 4, maxSize =10) identical(fgseaRes, fgseaRes2) # [1] FALSE R> fgseaRes[1:3, c("ES", "NES", "pval")] ES NES pval 1: -0.3087414 -0.6545067 0.8630705 2: 0.4209054 1.0360288 0.4312268 3: -0.4065043 -0.8617561 0.6431535 R> fgseaRes2[1:3, c("ES", "NES", "pval")] ES NES pval 1: 0.4554877 0.8284722 0.7201767 2: 0.5520914 1.1604366 0.2833333 3: -0.2908641 -0.5971951 0.9442724However, scaling won't change anything (because scaling does not change the directions?).
set.seed(42) fgseaRes3 <- fgsea(pathways = examplePathways, stats = 2*exampleRanks, minSize = 4, maxSize =10) identical(fgseaRes, fgseaRes3) # [1] TRUE - Obtain enrichment scores by calling fgsea(); i.e. fgseaMultilevel().
examplePathways[["5991601_Phosphorylation_of_Emi1"]] |> length() # [1] 8 set.seed(1) fgsea2 <- fgsea(pathways = examplePathways["5991601_Phosphorylation_of_Emi1"], stats = exampleRanks) tibble::as_tibble(fgsea2) # A tibble: 1 × 8 # pathway pval padj log2err ES NES size leadi…¹ # <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <list> # 1 5991601_Phosphor… 9.39e-7 9.39e-7 0.659 0.947 2.05 6 <chr> # … with abbreviated variable name ¹leadingEdge plotGseaTable(examplePathways["5991601_Phosphorylation_of_Emi1"], exampleRanks, fgsea2) - The computation speed is FAST!
system.time(invisible(fgsea(examplePathways, exampleRanks))) # user system elapsed # 31.162 38.427 15.800
Subramanian algorithm
In the plot, (x-axis) genes are sorted by their expression across all samples. Y-axis represents enrichment score. See HOW TO PERFORM GSEA - A tutorial on gene set enrichment analysis for RNA-seq. Bars represents genes being in the gene set. Genes on the LHS/RHS are more highly expressed in the experimental/control group. Small p means this gene set is enriched in this experimental sample.
Rafael A Irizarry
Gene set enrichment analysis made simple
RBiomirGS for microRNA/miRNA
- RBiomirGS R package
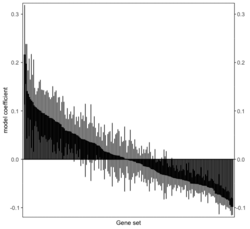
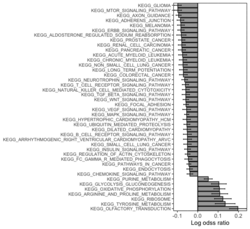
require("RBiomirGS") # two input files # liver.csv # kegg.v5.2.entrez.gmt, 186 pathways raw <- read.csv(file = "liver.csv", header = TRUE, stringsAsFactors = FALSE) dim(raw) # [1] 85 3 raw[1:3,] # miRNA FC pvalue # 1 mmu-miR-let-7f-5p 0.58 0.034 # 2 mmu-miR-1a-5p 0.32 0.037 # 3 mmu-miR-1b-5p 0.50 0.001 # Target mRNA mapping # creating 95 csv files; e.g. "mmu-miR-1a-5p_mRNA.csv" rbiomirgs_mrnascan(objTitle = "mmu_liver_predicted", mir = raw$miRNA, sp = "mmu", addhsaEntrez = TRUE, queryType = "predicted", parallelComputing = TRUE, clusterType = "PSOCK") # > head -3 mmu-miR-10b-5p_mRNA.csv # "database","mature_mirna_acc","mature_mirna_id","target_symbol","target_entrez","target_ensembl","score" # "diana_microt","MIMAT0000208","mmu-miR-10b-5p","Bdnf","12064","ENSMUSG00000048482","1" # "diana_microt","MIMAT0000208","mmu-miR-10b-5p","Bcl2l11","12125","ENSMUSG00000027381","1" # GSEA # generating 3 files: # mirnascore.csv # mrnascore.csv # mirna_mrna_iwls_GS.csv rbiomirgs_logistic(objTitle = "mirna_mrna_iwls", mirna_DE = raw, var_mirnaName = "miRNA", var_mirnaFC = "FC", var_mirnaP = "pvalue", mrnalist = mmu_liver_predicted_mrna_hsa_entrez_list, mrna_Weight = NULL, gs_file = "kegg.v5.2.entrez.gmt", optim_method = "IWLS", p.adj ="fdr", parallelComputing = FALSE, clusterType = "PSOCK") > wc -l mirnascore.csv 86 mirnascore.csv > head -3 mirnascore.csv 13:46:47 "miRNA","S_mirna" "mmu-miR-let-7f-5p",1.46852108295774 "mmu-miR-1a-5p",1.43179827593301 > wc -l mrnascore.csv 15297 mrnascore.csv > head -3 mrnascore.csv 13:46:53 "EntrezID","S_mrna" "1",-3.20388726317687 "10",-11.7197772358718 > wc -l mirna_mrna_iwls_GS.csv 187 mirna_mrna_iwls_GS.csv > head -3 mirna_mrna_iwls_GS.csv # "GS","converged","loss","gene.tested","coef","std.err","t.value","p.value","adj.p.val" # "KEGG_GLYCOLYSIS_GLUCONEOGENESIS","Y",0.0231448486023595,54,0.103936359331543,...# Histogram of all gene sets # y-axis = model coefficients # x-axis = gene set rbiomirgs_bar(gsadfm = mirna_mrna_iwls_GS, n = "all", y.rightside = TRUE, yTxtSize = 8, plotWidth = 300, plotHeight = 200, xLabel = "Gene set", yLabel = "model coefficient") # Histogram of top 50 ranked KEGG pathways based on absolute value of the coefficient # (with significant adjusted p values < 0.05) # y-axis = gene set # x-axis = log odds ratio rbiomirgs_bar(gsadfm = mirna_mrna_iwls_GS, gs.name = TRUE, n = 50, y.rightside = FALSE, yTxtSize = 8, plotWidth = 200, plotHeight = 300, xLabel = "Log odss ratio", signif_only = TRUE) # Volcano plot of all gene sets # y-axis = -log10(p-value) # x-axis = model coefficient rbiomirgs_volcano(gsadfm = mirna_mrna_iwls_GS, topgsLabel = TRUE, n =15, gsLabelSize = 3, sigColour = "blue", plotWidth = 250, plotHeight = 220, xLabel = "model coefficient")
 ,
,  ,
, 
- MicroRNAs (miRNAs) is a family of non-coding RNAs. miRNAs post-transcriptionally regulate the gene expression.
- miRBase: the microRNA database
- One microRNA/miR is one set. For human, its name (miRBase ID) looks like hsa-miR-181a, miRBase Accession Number is MIMAT0000256.
- MicroRNA and microRNA target database
- Correlation of expression profiles between microRNAs and mRNA targets using NCI-60 data
multiGSEA
multiGSEA - Combining GSEA-based pathway enrichment with multi omics data integration.
Plot
- Chapter 12 Visualization of Functional Enrichment Result, enrichplot::gseaplot() or enrichplot::gseaplot2() from Bioconductor
- GSEA 从数据到美图 where step4output.Rdata is available on github. Some quirks:
# geneset$term = str_remove(geneset$term,"HALLMARK_")
- fgsea::plotGseaTable() from Bioc
- limma:barcodeplot(), It is of course not the same as the Broad Institute's GSEA plot.
- 15 Visualization of functional enrichment result. Especially bar plots.
- Plot rank densities from singscore::plotRankDensity() from Bioc
Single sample
singscore
- singscore
- Paper Single sample scoring of molecular phenotypes. Our approach does not depend upon background samples and scores are thus stable regardless of the composition and number of samples being scored. In contrast, scores obtained by GSVA, z-score, PLAGE and ssGSEA can be unstable when less data are available (NS < 25). Simulation was conducted.
- SingscoreAMLMutations
- TotalScore = UpScore + DownScore
- centerScore and knownDirection(=TRUE by default) parameters used in generateNull() and simpleScore() functions.
- On the paper, epithelial and mesenchymal gene sets are up-regulated and TGFβ-EMT signature is bidirectional.
- The pvalue calculation seems wrong. For example, the first sample D_Ctrl_R1 returns p=0.996 but it should be 2*(1-.996) according to the null distribution plot. However, the 1st sample is a control sample so we don't expect it has a large score; its p-value should be large?
- Gene-set enrichment analysis workshop. Also use ExperimentHub, SummarizedExperiment, emtdata, GSEABase, msigdb, edgeR, limma, vissE, igraph & patchwork packages.
- Functions in the GSEABase package help with reading, parsing and processing the signatures.
ssGSEA & GSVA package
- https://github.com/broadinstitute/ssGSEA2.0
- ssGSEAProjection (v9.1.2). Each ssGSEA enrichment score represents the degree to which the genes in a particular gene set are coordinately up- or down-regulated within a sample.
- single sample GSEA (ssGSEA) from http://baderlab.org/
- How single sample GSEA works
- Data formats gct (expression data format), gmt (gene set database format).
- gmt file can be read by GSA.read.gmt()
- gmt file can be read by clusterProfiler::read.gmt(). See R做GSEA富集分析
- ssGSEA produced by Broad Gene Pattern is different from one produced by gsva package.
- Actually they are not compatible. See a plot in singscore paper.
- Gene Pattern also compute p-values testing the Spearman correlation
- GSVA and SSGSEA for RNA-Seq TPM data.
- Generally, negative enrichment values imply down-regulation of a signature / pathway; whereas, positive values imply up-regulation.
- The idea is to then conduct a differential signature / pathway analysis (using, for example, limma) so that you can have, in addition to differentially expressed genes, differentially expressed signatures / pathways.
- GSVA vignette. In GSEA Subramanian et al. (2005) it is also observed that the empirical null distribution obtained by permuting phenotypes is bimodal and, for this reason, significance is determined independently using the positive and negative sides of the null distribution.
- Tips:
- Pre-processing RNA-seq data (normalization and transformation) for GSVA
- pre-filtering gene sets for GSVA/ssGSEA
- kcdf parameter it is only relevant when method="gsva". See Method and kcdf arguments in gsva package and debugging source code.
- mx.diff: Offers two approaches to calculate the enrichment statistic (ES) from the KS random walk statistic. mx.diff=FALSE: ES is calculated as the maximum distance of the random walk from 0. mx.diff=TRUE (default): ES is calculated as the magnitude difference between the largest positive and negative random walk deviations.
- min.sz=1, max.sz=Inf
- 纯R代码实现ssGSEA算法评估肿瘤免疫浸润程度. The pheatmap package was used to draw the heatmap. Original paper.
- fpkm expression data was sorted by gene name & median values. Duplicated genes are removed. The data is transformed by log2(x+1) before sent to gsva()
- NES scores are scaled and truncated to [-2, 2]. The scores are further scaled to have the range [0,1] before sending it to the heatmap function
- gsva() from the GSVA package has an option to compute ssGSEA. 单样本基因集富集分析 --- ssGSEA (it includes the formula from Barbie's paper). The output of gsva() is a matrix of ES (# gene sets x # samples). It does not produce plots nor running the permutation tests. Notice the option ssgsea.norm. Note ssgsea.norm = TRUE (default) option will scale ES by the absolute difference of the max and min ES; Unscaled ES / (max(unscaled ES) - min(unscaled ES)). So the scaled ES values depends on the included samples. But it seems the impact of the included samples is small from some real data. See the source code on github and on how to debug an S4 function.
library(GSVA) library(heatmaply) p <- 10 ## number of genes n <- 30 ## number of samples nGrp1 <- 15 ## number of samples in group 1 nGrp2 <- n - nGrp1 ## number of samples in group 2 ## consider three disjoint gene sets geneSets <- list(set1=paste("g", 1:3, sep=""), set2=paste("g", 4:6, sep=""), set3=paste("g", 7:10, sep="")) ## sample data from a normal distribution with mean 0 and st.dev. 1 set.seed(1234) y <- matrix(rnorm(n*p), nrow=p, ncol=n, dimnames=list(paste("g", 1:p, sep="") , paste("s", 1:n, sep=""))) ## genes in set1 are expressed at higher levels in the last 'nGrp1+1' to 'n' samples y[geneSets$set1, (nGrp1+1):n] <- y[geneSets$set1, (nGrp1+1):n] + 2 gsva_es <- gsva(y, geneSets, method="ssgsea") dim(gsva_es) # 3 x 30 hist(gsva_es) # bi-modal range(gsva_es) # [1] -0.4390651 0.5609349 ## build design matrix design <- cbind(sampleGroup1=1, sampleGroup2vs1=c(rep(0, nGrp1), rep(1, nGrp2))) ## fit the same linear model now to the GSVA enrichment scores fit <- lmFit(gsva_es, design) ## estimate moderated t-statistics fit <- eBayes(fit) ## set1 is differentially expressed topTable(fit, coef="sampleGroup2vs1") # logFC AveExpr t P.Value adj.P.Val B # set1 0.5045008 0.272674410 8.065096 4.493289e-12 1.347987e-11 17.067380 # set2 -0.1474396 0.029578749 -2.315576 2.301957e-02 3.452935e-02 -4.461466 # set3 -0.1266808 0.001380826 -2.060323 4.246231e-02 4.246231e-02 -4.992049 heatmaply(gsva_es) # easy to see a pattern # samples' clusters are not perfect heatmaply(gsva_es, scale = "none") # 'scale' is not working? heatmaply(y, Colv = F, Rowv= F, scale = "none") # not easy to see a pattern - If we set ssgsea.norm=FALSE, do we get the same results when we compute ssgsea using a subset of samples?
gsva1 <- gsva(y, geneSets, method="ssgsea", ssgsea.norm = FALSE) gsva2 <- gsva(y[, 1:2], geneSets, method="ssgsea", ssgsea.norm = FALSE) range(abs(gsva1[, 1:2] - gsva2)) # [1] 0 0
- Verify ssgsea.norm = TRUE option.
gsva_es <- gsva(y, geneSets, method="ssgsea") gsva1 <- gsva(y, geneSets, method="ssgsea", ssgsea.norm = FALSE) gsva2 <- gsva1 / abs(max(gsva1) - min(gsva1)) # abs(max(gsva1) - min(gsva1)) 8.9 range(gsva_es) # [1] -0.4390651 0.5609349 range(abs(gsva_es - gsva2)) # [1] 0 0
- Only ranks matters! If I replace the sample 1 gene expression values with the ranks, the ssgsea scores are not changed at all.
y2 <- y y2[, 1] <- rank(y2[, 1]) gsva2 <- gsva(y2, geneSets, method="ssgsea") gsva_es[, 1] # set1 set2 set3 # 0.1927056 0.1699799 -0.1782934 gsva2[, 1] # set1 set2 set3 # 0.1927056 0.1699799 -0.1782934 all.equal(gsva_es, gsva2) # [1] TRUE # How about the reverse ranking? No that will change everything. # The one with the smallest value was assigned one according to # the definition of 'rank' y3[, 1] <- 11 - y2[, 1] gsva3 <- gsva(y3, geneSets, method="ssgsea") gsva3[, 1] # set1 set2 set3 #-0.054991319 -0.009603802 0.191733634 cbind(y[, 1], y2[, 1], y3[, 1]) # [,1] [,2] [,3] # g1 -1.2070657 2 9 # g2 0.2774292 7 4 # g3 1.0844412 10 1 # g4 -2.3456977 1 10 # g5 0.4291247 8 3 # g6 0.5060559 9 2 # g7 -0.5747400 4 7 # g8 -0.5466319 6 5 # g9 -0.5644520 5 6 # g10 -0.8900378 3 8
- Is mx.diff useful? No.
gsva4 <- gsva(y, geneSets, method="ssgsea", mx.diff = FALSE) all.equal(gsva_es, gsva4) # [1] TRUE
- A simple implementation of ssGSEA (single sample gene set enrichment analysis)
- Use "ssgsea-gui.R". The first question is a folder containing input files GCT. The 2nd question is about gene set database in GMT format. This has to be very restrict. For example, "ptm.sig.db.all.uniprot.human.v1.9.0.gmt" and "ptm.sig.db.all.sitegrpid.human.v1.9.0.gmt" provided in github won't work with the example GCT file.
setwd("~/github/ssGSEA2.0/") source("ssgsea-gui.R") # select a folder containing gct files; e.g. PI3K_pert_logP_n2x23936.gct # select a gene set file; e.g. <ptm.sig.db.all.flanking.human.v1.8.1.gmt>A new folder (e.g. 2021-03-01) will be created under the same parent folder as the gct file folder.
tree -L 1 ~/github/ssGSEA2.0/example/gct/2021-03-20/ ├── PI3K_pert_logP_n2x23936_ssGSEA-combined.gct ├── PI3K_pert_logP_n2x23936_ssGSEA-fdr-pvalues.gct ├── PI3K_pert_logP_n2x23936_ssGSEA-pvalues.gct ├── PI3K_pert_logP_n2x23936_ssGSEA-scores.gct ├── PI3K_pert_logP_n2x23936_ssGSEA.RData ├── parameters.txt ├── rank-plots ├── run.log └── signature_gct tree ~/github/ssGSEA2.0/example/gct/2021-03-20/rank-plots | head -3 # 102 files. One file per matched gene set ├── DISEASE.PSP_Alzheime_2.pdf ├── DISEASE.PSP_breast_c_2.pdf tree ~/github/ssGSEA2.0/example/gct/2021-03-20/signature_gct | head -3 # 102 files. One file per matched gene set ├── DISEASE.PSP_Alzheimer.s_disease_n2x23.gct ├── DISEASE.PSP_breast_cancer_n2x14.gct
Since the GCT file contains 2 samples (the last 2 columns), ssGSEA produces one rank plot for each gene set (with adjusted p-value & the plot could contain multiple samples). The ES scores are saved in <PI3K_pert_logP_n2x23936_ssGSEA-scores> and adjust p-values are saved in <PI3K_pert_logP_n2x23936_ssGSEA-fdr-pvalues>.
- Some possible problems using ssGSEA to replace GSEA for DE analysis? See a toy example on Single Sample GSEA (ssGSEA) and dynamic range of expression. PS: the rank values table is wrong; they should be reversed.
- In general terms, PLAGE and z-score are parametric and should perform well with close-to-Gaussian expression profiles, and ssGSEA and GSVA are non-parametric and more robust to departures of Gaussianity in gene expression data. See Method and kcdf arguments in gsva package
- Some discussions from biostars.org. Find -> "ssgsea"
- Some papers.
- 【生信分析 3】教你看懂GSEA和ssGSEA分析结果. No groups/classes in the data (6:33). Output is a heatmap. Each value is computed sample by sample. Rows = gene set. Columns = (sorted by the 1st gene set) samples.
escape
escape - Easy single cell analysis platform for enrichment. Github.
Benchmark
Toward a gold standard for benchmarking gene set enrichment analysis Geistlinger, 2021. GSEABenchmarkeR package in Bioconductor.
phantasus
- phantasus Visual and interactive gene expression analysis. read.gct(), write.gct(), getGSE(), loadGEO(), gseaPlot().
- Using Phantasus application
PhenoExam
PhenoExam: gene set analyses through integration of different phenotype databa
Selected papers
- On the influence of several factors on pathway enrichment analysis Mubeen 2022
- Popularity and performance of bioinformatics software: the case of gene set analysis Xie 2021
- Gene set analysis approaches for RNA-seq data: performance evaluation and application guideline Rahmatallah 2015. Simulated RNA-Seq data was considered too.
- Gene set analysis methods: a systematic comparison Mathur, 2018
- A Comparison of Gene Set Analysis Methods in Terms of Sensitivity, Prioritization and Specificity Tarca 2013
Malacards
- MalaCards - human disease database. Paper 2018
- GSEABenchmarkeR package used it.
- (Not Malacards) KEGGandMetacoreDzPathwaysGEO Disease Datasets from GEO. This is a collection of 18 data sets for which the phenotype is a disease with a corresponding pathway in either KEGG or metacore database.This collection of datasets were used as gold standard in comparing gene set analysis methods.
- 生信数据库之疾病数据库
GSDA
Gene-set distance analysis (GSDA): a powerful tool for gene-set association analysis
MSigDB
https://www.gsea-msigdb.org/gsea/msigdb/
- C1: Hallmark
- C2: Curated gene sets including BioCarta, KEGG, Reactome
- C5: Oncology
Hallmark
- HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION/EMT from MsigDB
- Epithelial (170 genes), mesenchymal gene signatures 218-170 genes). Tan et al. 2014 and can be found in the ‘Table S1B. Generic EMT signature for cell line’ (Epi/Mes column) in the supplementary tables file. See here. I save the gene lists in Github.