R packages: Difference between revisions
| (227 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
= R package management = | = R package management = | ||
== Challenges in Package Management == | |||
[https://www.rstudio.com/blog/challenges-in-package-management/ Challenges in Package Management] | |||
== Packages loaded at startup == | == Packages loaded at startup == | ||
''getOption("defaultPackages")'' | ''getOption("defaultPackages")'' | ||
== How to install a new package == | == How to install a new package == | ||
* [https://thomasadventure.blog/posts/install-r-packages/ The Comprehensive Guide to Installing R Packages from CRAN, Bioconductor, GitHub and Co.] | |||
* An example from [https://bvieth.github.io/powsimR/articles/powsimR.html powsimR] package | |||
=== utils::install.packages() === | === utils::install.packages() === | ||
* By default, install.packages() will check versions and install uninstalled packages shown in ''' 'Depends', 'Imports' ''', and ''' 'LinkingTo' ''' fields. See [http://cran.r-project.org/doc/manuals/r-release/R-exts.html R-exts] manual. | * By default, install.packages() will check versions and install uninstalled packages shown in ''' 'Depends', 'Imports' ''', and ''' 'LinkingTo' ''' (<span style="color: red">DIL</span>) fields. See [http://cran.r-project.org/doc/manuals/r-release/R-exts.html R-exts] manual. | ||
* Take advantage of '''Ncpus''' parameter in [https://www.rdocumentation.org/packages/utils/versions/3.6.2/topics/install.packages install.packages()] | * Take advantage of '''Ncpus''' parameter in [https://www.rdocumentation.org/packages/utils/versions/3.6.2/topics/install.packages install.packages()] | ||
* If we want to install packages listed in 'Suggests' field, we should specify it explicitly by using ''dependencies'' argument: | * By default dependencies = NA, which means c("Depends", "Imports", "LinkingTo"). | ||
: <syntaxhighlight lang=' | * If we want to install packages listed in '''Suggests''' (or 'Enhances') field, we should specify it explicitly by using ''dependencies'' argument: | ||
: <syntaxhighlight lang='r'> | |||
install.packages(XXXX, dependencies = c("Depends", "Imports", "Suggests", "LinkingTo")) | install.packages(XXXX, dependencies = c("Depends", "Imports", "Suggests", "LinkingTo")) | ||
# OR | # OR | ||
| Line 39: | Line 46: | ||
: Look at CRAN, we see the latest version (1.7-0) of [https://cran.r-project.org/web/packages/gridSVG/index.html gridSVG] depends on the latest version of R (3.6.0). But '''why even the source is removed from the old version of R?''' | : Look at CRAN, we see the latest version (1.7-0) of [https://cran.r-project.org/web/packages/gridSVG/index.html gridSVG] depends on the latest version of R (3.6.0). But '''why even the source is removed from the old version of R?''' | ||
* The '''install.packages''' function source code can be found in R -> src -> library -> utils -> R -> [https://github.com/wch/r-source/blob/trunk/src/library/utils/R/packages2.R packages2.R] file from [https://github.com/wch/r-source Github] repository (put 'install.packages' in the search box). | * The '''install.packages''' function source code can be found in R -> src -> library -> utils -> R -> [https://github.com/wch/r-source/blob/trunk/src/library/utils/R/packages2.R packages2.R] file from [https://github.com/wch/r-source Github] repository (put 'install.packages' in the search box). | ||
=== Change to use curl === | |||
Use '''options(download.file.method = "curl")''' before calling install.packages() or other install methods. | |||
The default on Linux/MacOS is "auto" -> usually "wget" or "libcurl" if available, else "internal". When we force R to call the system's '''curl''' binary directly, bypassing wget or R's internal downloaded. Sometimes "wget" doesn't handle HTTPS well if SSL libraries aren't set up right. '''curl often works better with HTTPS and proxies if properly installed'''. | |||
=== mirror === | === mirror === | ||
| Line 72: | Line 84: | ||
pacman::p_load("BioManager") | pacman::p_load("BioManager") | ||
pacman::p_load("DESeq2",try.bioconductor=TRUE,update.bioconductor=TRUE) | pacman::p_load("DESeq2",try.bioconductor=TRUE,update.bioconductor=TRUE) | ||
</pre> | |||
PS. Brute force method | |||
<pre> | |||
mypackages<-c("ggplot2", "dplyr") | |||
for (p in mypackages){ | |||
cond <- suppressWarnings(!require(p, character.only = TRUE)) | |||
if(cond){ | |||
try(install.packages(p), silent = TRUE) | |||
library(p, character.only = TRUE) | |||
} | |||
} | |||
</pre> | </pre> | ||
| Line 81: | Line 106: | ||
* https://cran.r-project.org/web/packages/remotes/index.html | * https://cran.r-project.org/web/packages/remotes/index.html | ||
* https://www.rdocumentation.org/packages/remotes/versions/2.0.2 | * https://www.rdocumentation.org/packages/remotes/versions/2.0.2 | ||
* remotes::install("USERNAME/REPOSITORY"). [https://twitter.com/strnr/status/1781419059948462394 Install all my packages with just a github repository and a DESCRIPTION file] | |||
To install a package from a local machines with all dependency, run '''remotes::install_local(path = "MyPackage.tar.gz", dependencies=TRUE)''' or '''devtools::install()''' though the later requires to untar the source first. | To install a package from a local machines with all dependency, run '''remotes::install_local(path = "MyPackage.tar.gz", dependencies=TRUE)''' or '''devtools::install()''' though the later requires to untar the source first. | ||
==== remotes::install_github() vs devtools::install_github() ==== | |||
See [https://twitter.com/JohnHelveston/status/1417798263344930819 the answers]. '''remotes''' is a lighter weight package for those who don't need/want all the other functionality in devtools. The remotes package says it copied most of its functionality from devtools, so the functions are likely the same. | |||
=== devtools === | === devtools === | ||
[https://www.rdocumentation.org/packages/devtools/versions/2.0.1/topics/install devtools::install()] | * [https://www.rdocumentation.org/packages/devtools/versions/2.0.1/topics/install devtools::install()] | ||
* devtools::install(".", ...) assumes the current location contains the package content; eg [https://github.com/seandavi/BuildABiocWorkshop2020/blob/master/Dockerfile Dockerfile] from [https://github.com/seandavi/BuildABiocWorkshop2020 Bioc2020 template]. | |||
=== pak === | === pak === | ||
https://cran.r-project.org/web/packages/pak/index.html | * https://cran.r-project.org/web/packages/pak/index.html. 'pak' supports CRAN, 'Bioconductor' and 'GitHub' packages. An example about the [https://tidymodels.github.io/censored/index.html censored] package. | ||
* When I use rsconnect::deployApp() to deploy a shiny app to shinyapps.io, the terminal emits the message ''The following package(s) were installed from sources, but may be available from the following remotes: - pak [r-lib/pak]''. So it seems shinyapps.io uses the [https://github.com/r-lib/pak pak] to install packages. | |||
* [https://www.r-bloggers.com/2023/08/three-four-r-functions-i-enjoyed-this-week/ How does this package depend on this other package?] | |||
=== BiocManager === | === BiocManager === | ||
| Line 100: | Line 132: | ||
=== Ubuntu === | === Ubuntu === | ||
<ul> | |||
<li>[http://dirk.eddelbuettel.com/blog/2017/12/22/ Finding Binary .deb Files for CRAN Packages] | |||
<li>Add an additional binary source to your source.lists file, see for example, the line in [https://github.com/rocker-org/rocker/blob/master/r-ubuntu/Dockerfile#L27 rocker]. This is mentioned in [https://www.jumpingrivers.com/blog/faster-r-package-installation-rstudio/ Faster R package installation]. | |||
<li>[http://dirk.eddelbuettel.com/blog/2020/06/22/#027_ubuntu_binaries R and CRAN Binaries for Ubuntu] by Dirk Eddelbuettel | |||
* [https://eddelbuettel.github.io/r2u/ r2u: R Binaries for Ubuntu]; see [https://twitter.com/eddelbuettel/status/1523376755548979201 The new #CRANapt repo has 19000 .deb binaries (and 200+ from BioConductor) for each with full dependencies and `apt` integration.] | |||
[https://www. | ** [https://www.brodrigues.co/blog/2022-10-29-mkusb_minp/ A Linux Live USB as a statistical programming dev environment] | ||
** [https://yegrug.github.io/meetups/2022-10-27/ October 27, 2022: r2u, binary packages on Ubuntu] | |||
* [https://enchufa2.github.io/rspm/ rspm]. [https://www.enchufa2.es/archives/rspm-easy-access-to-rspm-binary-packages-with-automatic-management-of-system-requirements.html {rspm}: easy access to RSPM binary packages with automatic management of system requirements] | |||
* [https://cloud.r-project.org/web/packages/bspm/index.html bspm] R package (cf '''rocker/r-rspm''' docker image which is not an R package). 5 lines. After that, "install.packages()" will trigger "sudo apt-get install". Note: it seems the ''apt-add-repository'' commands will break the ''apt update'' command on '''Debian 11'''. After a successful installation on Ubuntu OS, I'll see the following messages every time I started R, | |||
<pre> | <pre> | ||
install.packages | Loading required package: utils | ||
Tracing function "install.packages" in package "utils" | |||
</pre> | </pre> | ||
Also [https://github.com/Enchufa2/bspm/issues/43 for some reason], I cannot remove packages that were just installed. Not sure if the same problem happened if I use '''rig''' (based on rspm). | |||
<pre> | <pre> | ||
> install.packages("fgsea") # Works fine | |||
> remove.packages("fgsea") | |||
> | Removing package from ‘/home/brb/R/x86_64-pc-linux-gnu-library/4.2’ | ||
(as ‘lib’ is unspecified) | (as ‘lib’ is unspecified) | ||
Error in find.package(pkgs, lib) : there is no package called ‘fgsea’ | |||
> packageVersion("fgsea") | |||
> install.packages(" | [1] ‘1.22.0’ | ||
... | </pre> | ||
* | Since the ''bspm::enable()'' function enables the integration of '''install_sys()''' into '''install.packages()''', we have to use '''remove_sys(pkgs)''' to uninstall a package. See [https://rdrr.io/cran/bspm/api/ API]. | ||
</li> | |||
</ul> | |||
=== Posit Package Manager/RStudio Package Manager/PPM, Posit Public Package Manager/P3M === | |||
<ul> | |||
<li>https://packagemanager.posit.co/client/#/ -> Repository:Bioconductor -> Setup. | |||
* On Ubuntu (Linux), there is no binary format of Bioconductor packages. P3M only has binary format of CRAN packages. | |||
* On Windows, I have to install '''Rtools'''. After Rtools has been installed, R will recognize it. Go to https://packagemanager.posit.co/client/#/. Select Bioconductor and click "SETUP". Pick Bioconductor version. Copy/paste options() statements into R or put it in the '''~/.Rprofile'''. | |||
* On macOS, similar to Windows. | |||
<li>[https://packagemanager.rstudio.com/__docs__/admin/getting-started/installation/installing-ppm/ PPM Admin Guide]. <s>It only supports Ubuntu (no Debian), RedHat, OpenSUSE.</s> | |||
* In the section [https://docs.posit.co/rspm/admin/serving-binaries/#binary-availability CRAN Binary Availability], it says ''Some common reasons why a package may not have a binary are: A package depends on a Bioconductor package. Package Manager does not currently support binary packages from Bioconductor, notable packages include Seurat and WGCNA.'' | |||
* [https://docs.posit.co/faq/faq-files/p3m-faq.html Posit Public Package Manager FAQ] | |||
<li>[https://docs.posit.co/rspm/admin/serving-binaries/#installing-binary-packages R Configuration Steps (Linux) -> Using Linux Binary Packages -> Configuring the R User Agent Header] from Admin Guide. <span style="color: red">The step of configuring http user agent header should not be skipped</span>. | |||
<li>[https://tshafer.com/blog/2023/07/posit-package-manager-linux Posit Package Manager for Linux R Binaries]. Put the following in the '''~/.Rprofile''' file on my Debian OS. If you use the RStudio IDE, the software automatically injects this User-Agent string behind the scenes when R starts. | |||
> | <syntaxhighlight lang='r'> | ||
[ | options(HTTPUserAgent = sprintf( | ||
> options() | "R/%s R (%s)", | ||
getRversion(), | |||
" | paste( | ||
getRversion(), | |||
R.version["platform"], | |||
R.version["arch"], | |||
R.version["os"] | |||
) | |||
)) | |||
== | # Configure BioCManager to use Posit Package Manager: | ||
options(BioC_mirror = "https://packagemanager.posit.co/bioconductor") | |||
options(BIOCONDUCTOR_CONFIG_FILE = "https://packagemanager.posit.co/bioconductor/config.yaml") | |||
local({ | |||
# 1. Initialize with a default CRAN mirror (fallback) | |||
r <- getOption("repos") | |||
r["CRAN"] <- "https://cloud.r-project.org" | |||
# 2. Check if we are on Linux and can read os-release | |||
if (Sys.info()["sysname"] == "Linux" && file.exists("/etc/os-release")) { | |||
# Read the OS release file | |||
os_info <- readLines("/etc/os-release", warn = FALSE) | |||
# Extract the codename (e.g., "bookworm", "bullseye", "jammy") | |||
# Looks for line: VERSION_CODENAME=bookworm | |||
codename_line <- grep("^VERSION_CODENAME=", os_info, value = TRUE) | |||
if (length(codename_line) > 0) { | |||
# Remove the key and any quotes | |||
codename <- sub("^VERSION_CODENAME=\"?([^\"]+)\"?.*$", "\\1", codename_line) | |||
# 3. Construct the Posit Package Manager URL | |||
# Note: This assumes you want the 'latest' snapshot. | |||
ppm_url <- paste0("https://packagemanager.posit.co/cran/__linux__/", codename, "/latest") | |||
# Set the repo | |||
r["CRAN"] <- ppm_url | |||
} | |||
} | |||
# Apply the options | |||
options(repos = r) | |||
}) | |||
# Verify | |||
# getOption("repos") | |||
</syntaxhighlight> | </syntaxhighlight> | ||
<li>It provides '''pre-compiled binary packages''' for Linux and Windows OS, but not macOS. When enabled, RStudio Package Manager will serve the appropriate CRAN binaries to R users instead of the source packages, saving R users significant installation time. | |||
* <strike>It does not include binary packages from bioconductor</strike>. However, Package Manager allows you to supplement local and git sources with your own precompiled binaries. | |||
* [https://support.posit.co/hc/en-us/articles/360016400354-Using-Bioconductor-with-RStudio-Package-Manager Using Bioconductor with RStudio Package Manager] 7/12/2021 | |||
* See [https://docs.posit.co/rspm/admin/serving-binaries/ Serving Package Binaries] | |||
<li>It is similar to MRAN as it allows users to use particular a '''snapshot''' of CRAN as their active repositories within their R session | |||
* [https://community.rstudio.com/t/programmatically-retrieve-frozen-urls-for-cran-from-packagemanager-rstudio-com/119326 programmatically retrieve frozen urls for CRAN from packagemanager.rstudio.com] | |||
* See [https://packagemanager.rstudio.com/client/#/repos/2/overview Setup]. We can change the distribution (Source, CentOS7, Rocky Linux 9, OpenSUSE, Red Hat, SLES, Ubuntu, Windows, macOS) for the Repository URL. | |||
* See [https://packagemanager.rstudio.com/__docs__/user/ User Guide] | |||
<li> Once configured, users can access RStudio Package Manager through all their familiar tools including base R’s install.packages(), packrat, and devtools. | |||
<li>[https://blog.rstudio.com/2020/07/01/announcing-public-package-manager/ Announcing Public Package Manager and v1.1.6] | |||
<li>Package Manager allows linux users to install '''pre-built binaries''' of the packages which will make install quicker. See an example [https://blog.sellorm.com/2021/04/25/shiny-app-in-docker/?s=09 here]. | |||
<pre> | |||
# Freeze to Apr 29, 2021 8:00 PM | |||
options(repos = c(REPO_NAME = "https://packagemanager.posit.co/all/2639103")) | |||
# Using Linux Binary Packages | |||
https://r-pkgs.example.com/our-cran/__linux__/xenial/latest | |||
https://packagemanager.posit.co/cran/__linux__/focal/2021-04-23 | |||
</pre> | </pre> | ||
<li>[https://www.rstudio.com/blog/publishing-your-own-binary-packages-with-rspm-2022-07/ Bring Your Own Binary Packages with RSPM] | |||
<li>'''Docker images''' | |||
* r-base, rocker/r-base don't use PPM (as of R 4.3.3). | |||
* rocker/r-ver (eg. rocker/r-ver:4.3.3) uses PPM. See the table of images on [https://rocker-project.org/images/ The versioned stack]. | |||
* [https://bioconductor.org/help/docker/ Bioconductor] (source on [https://github.com/Bioconductor/bioconductor_docker github]) does take advantage of PPM. In Bioc 3.17/R 4.3.1, Posit's PPM hosts the binaries. As of Bioc 3.18/R 4.3.3, Bioconductor hosts the binaries.</br> | |||
:RStudio will be available on your web browser at http://localhost:8787. The username is "rstudio". | |||
:<syntaxhighlight lang='r'>docker run \ | |||
-e PASSWORD=bioc \ | |||
-p 8787:8787 \ | |||
bioconductor/bioconductor_docker:devel | |||
</syntaxhighlight> | |||
:R will be available on the terminal | |||
:<syntaxhighlight lang='r'> | |||
docker run -it --rm \ | |||
--user rstudio \ | |||
bioconductor/bioconductor_docker:RELEASE_3_18 R | |||
</syntaxhighlight> | |||
{{Pre}} | {{Pre}} | ||
system.time(BiocManager::install("DESeq2", ask = FALSE)) | |||
# Replacement repositories: | |||
# CRAN: https://p3m.dev/cran/__linux__/jammy/latest | |||
# ... | |||
# trying URL 'https://bioconductor.org/packages/3.18/container-binaries/bioconductor_docker/src/contrib/bitops_1.0-7_R_x86_64-pc-linux-gnu.tar.gz' | |||
# | Content type 'application/gzip' length 24831 bytes (24 KB) | ||
# ... | |||
# 52 seconds | |||
</pre> | |||
</ul> | |||
install. | === install a package as it existed on a specific date/snapshot: mran repository === | ||
* [https://blog.revolutionanalytics.com/2023/01/mran-time-machine-retired.html MRAN Time Machine will be retired on July 1 2023] | |||
** [https://www.brodrigues.co/blog/2023-01-12-repro_r/ MRAN is getting shutdown] - what else is there for reproducibility with R, or why reproducibility is on a continuum? | |||
* [https://www.r-bloggers.com/mran-snapshots-and-you-2/ MRAN snapshots, and you] May 22, 2019. | |||
Look at https://cran.microsoft.com/snapshot/, it seems the snapshot went back 2014 for R 3.1.1. | |||
For example, | |||
<pre> | |||
options(repos = c(CRAN = "https://mran.microsoft.com/snapshot/2022-10-01")) | |||
install.packages("xgboost", repos="https://mran.microsoft.com/snapshot/2022-10-01/") | |||
</pre> | |||
This trick works great when I try to install an R package (glmnet 3.0-2) on an old version of R. The current glmnet requires R 3.6.0. ''So even the source package is not available for older versions of R.'' If I install a package that does not require a very new version of R, that it works. The same problem happens on Windows OS. (today is 2020-05-05) | |||
<pre> | |||
$ docker run --rm -it r-base:3.5.3 | |||
... | |||
> install.packages("glmnet") | |||
Installing package into ‘/usr/local/lib/R/site-library’ | |||
(as ‘lib’ is unspecified) | |||
Warning message: | |||
package ‘glmnet’ is not available (for R version 3.5.3) | |||
> install.packages("glmnet", repos="https://mran.microsoft.com/snapshot/2019-09-20/") | |||
... | |||
* DONE (glmnet) | |||
The downloaded source packages are in | |||
‘/tmp/RtmpoWVWFQ/downloaded_packages’ | |||
> packageVersion("glmnet") | |||
[1] ‘2.0.18’ | |||
> options()$repos | |||
CRAN | |||
"https://cloud.r-project.org" | |||
> install.packages('knitr') # OK. | |||
</pre> | |||
== | === install a package on an old version of R === | ||
< | <ul> | ||
<li>Currently R is 4.2.2 | |||
< | <li>I have build R 4.1.3 and R 4.0.5 from source on Ubuntu 22.04. P.S. We need to include/enable "Source code" URIs [https://askubuntu.com/a/857433 Error :: You must put some 'source' URIs in your sources.list]. | ||
<pre> | |||
sudo apt update | |||
sudo apt-get build-dep r-base | |||
tar -xzvf R-4.1.3.tar.gz | |||
cd R-4.1.3 | |||
./configure | |||
time make -j2 | |||
bin/R | |||
< | </pre> | ||
<li>I'm trying to install DESEq2 on R 4.1.3 and R 4.0.5 | |||
<li>On R 4.1.3, the installation is successful. | |||
<pre> | <pre> | ||
install.packages("BiocManager") | |||
BiocManager::install("DESeq2") | |||
packageVersion("DESeq2") # 1.34.0 | |||
</pre> | </pre> | ||
<li>On R 4.0.5, the installation of "locfit" (current version 1.5-9.7) requires R >= 4.1.0. Check on CRAN, we see the locfit version is 1.5-6 for R 4.0.5 (2021-3-31). So I try this version first. | |||
<pre> | |||
install.packages("BiocManager") | |||
install.packages("remotes") | |||
remotes::install_version("locfit", "1.5-6") | |||
# Error: a 'NAMESPACE' file is required | |||
# | # Try the next version 1.5-7 | ||
remotes::install_version("locfit", "1.5-7") # Works | |||
BiocManager::install("DESeq2") # Works too | |||
packageVersion("DESeq2") # 1.30.1 | |||
</pre> | </pre> | ||
<li>Now it is a good opportunity to try '''renv'''. So I create a new folder 'testProject' and create a new file 'test.R'. This R file contains only one line: library(DESeq2). | |||
<pre> | |||
> setwd("testProj") | |||
> install.packages("renv") | |||
> renv::init() | |||
* Initializing project ... | |||
* Discovering package dependencies ... Done! | |||
* Copying packages into the cache ... [79/79] Done! | |||
One or more Bioconductor packages are used in your project, | |||
but the BiocManager package is not available. | |||
Consider installing BiocManager before snapshot. | |||
The following package(s) will be updated in the lockfile: | |||
... | |||
The version of R recorded in the lockfile will be updated: | |||
- R [*] -> [4.0.5] | |||
* Lockfile written to '~/Downloads/testProject/renv.lock'. | |||
* Project '~/Downloads/testProject' loaded. [renv 0.16.0] | |||
* renv activated -- please restart the R session. | |||
> renv::install("BiocManager") # NOT SURE IF THIS IS NECESSARY | |||
Retrieving 'https://cloud.r-project.org/src/contrib/BiocManager_1.30.19.tar.gz' ... | |||
OK [file is up to date] | |||
Installing BiocManager [1.30.19] ... | |||
OK [built from source] | |||
Moving BiocManager [1.30.19] into the cache ... | |||
OK [moved to cache in 20 milliseconds] | |||
> q() | |||
</pre> | |||
Now DESeq2 is in "renv.lock" file! | |||
Note if we accidentally start a different version of R on an renv-ed project based on an older version of R, we shall see some ''special'' message informing us '''The project library is out of sync with the lockfile'''. At this time, the "renv.lock" is not touched yet. | |||
<pre> | |||
# Bootstrapping renv 0.16.0 -------------------------------------------------- | |||
* Downloading renv 0.16.0 ... OK (downloaded source) | |||
* Installing renv 0.16.0 ... Done! | |||
* Successfully installed and loaded renv 0.16.0. | |||
! Using R 4.2.2 (lockfile was generated with R 4.0.5) | |||
* Project '~/Downloads/testProject' loaded. [renv 0.16.0] | |||
* The project library is out of sync with the lockfile. | |||
* Use `renv::restore()` to install packages recorded in the lockfile. | |||
</pre> | </pre> | ||
<li>What happened if we start with the same version of R but have a different version of renv in the global environment (vs renv.lock)? | |||
<pre> | |||
install.packages("remotes") | |||
remotes::install_version("renv", "0.15.5") | |||
renv::init() | |||
q() | |||
# Start the same R from another directory and update renv to the latest version 0.16.0 | |||
# Now go back to the project directory and start R to see what will happen? | |||
# Nothing.... This is because the same project has the old & same version of "renv" library | |||
# in the "$PROJECT/renv" directory | |||
</pre> | |||
If we delete the "renv" subdirectory, ".Rprofile" file but keep "renv.lock" file, and start R (remember the 'renv' package version in the global R is different from the version in "renv.lock". | |||
<pre> | |||
> renv::restore() | |||
This project has not yet been activated. | |||
Activating this project will ensure the project library is used during restore. | |||
Please see `?renv::activate` for more details. | |||
Would you like to activate this project before restore? [Y/n]: y | |||
Retrieving 'https://cloud.r-project.org/src/contrib/BiocManager_1.30.19.tar.gz' ... | |||
OK [file is up to date] | |||
Installing BiocManager [1.30.19] ... | |||
OK [built from source] | |||
Moving BiocManager [1.30.19] into the cache ... | |||
OK [moved to cache in 27 milliseconds] | |||
* Project '~/Downloads/testProject' loaded. [renv 0.16.0] | |||
* The project may be out of sync -- use `renv::status()` for more details. | |||
The following package(s) will be updated: | |||
=== | # Bioconductor ======================= | ||
- AnnotationDbi [* -> 1.56.2] | |||
... | |||
# CRAN =============================== | |||
- renv [0.16.0 -> 0.15.5] | |||
... | |||
Do you want to proceed? [y/N]: y | |||
Retrieving 'https://cloud.r-project.org/src/contrib/Archive/renv/renv_0.15.5.tar.gz' ... | |||
OK [downloaded 961.3 Kb in 0.2 secs] | |||
Installing renv [0.15.5] ... | |||
OK [built from source] | |||
Moving renv [0.15.5] into the cache ... | |||
OK [moved to cache in 6.8 milliseconds] | |||
... | |||
The following package(s) have been updated: | |||
renv [installed version 0.15.5 != loaded version 0.16.0] | |||
Consider restarting the R session and loading the newly-installed packages. | |||
</pre> | </pre> | ||
All the R packages are restored in the $PROJECT/renv directory. | |||
</li> | |||
</ul> | |||
== | == pkgbuild: Locates compilers == | ||
https:// | https://cran.r-project.org/web/packages/pkgbuild/index.html Find Tools Needed to Build R Packages | ||
<pre> | <pre> | ||
pkgbuild::check_build_tools(debug = TRUE) | |||
</pre> | |||
== Check installed Bioconductor version == | |||
Following [https://www.biostars.org/p/150920/ this post], use '''tools:::.BioC_version_associated_with_R_version()'''. | |||
''Mind the '.' in front of the 'BioC'. It may be possible for some installed packages to have been sourced from a different BioC version.'' | |||
<syntaxhighlight lang='rsplus'> | |||
</ | tools:::.BioC_version_associated_with_R_version() # `3.6' | ||
tools:::.BioC_version_associated_with_R_version() == '3.6' # TRUE | |||
</syntaxhighlight> | |||
== CRAN Package Depends on Bioconductor Package == | |||
For example, if I run ''install.packages("NanoStringNorm")'' to install the [https://cran.r-project.org/web/packages/NanoStringNorm/index.html package] from CRAN, I may get | |||
<pre> | <pre> | ||
ERROR: dependency ‘vsn’ is not available for package ‘NanoStringNorm’ | |||
</pre> | </pre> | ||
This is because the NanoStringNorm package depends on the vsn package which is on Bioconductor. | |||
Another example is CRAN's ''tidyHeatmap'' that imports ''ComplexHeatmap'' in Bioconductor. | |||
Another instance is CRAN's ''biospear'' (actually [https://cran.r-project.org/web/packages/plsRcox/index.html plsRcox]) that depends on Bioc's 'survcomp' & 'mixOmics'. | |||
One solution is to run a line '''setRepositories(ind=1:2)'''. See [http://stackoverflow.com/questions/14343817/cran-package-depends-on-bioconductor-package-installing-error this post] or [https://stackoverflow.com/questions/34617306/r-package-with-cran-and-bioconductor-dependencies this one]. Note that the default repository list can be found at (Ubuntu) '''/usr/lib/R/etc/repositories''' file. | |||
{{Pre}} | {{Pre}} | ||
options("repos") # display the available repositories (only CRAN) | |||
setRepositories(ind=1:2) | |||
"https://cran.rstudio.com/" | options("repos") # CRAN and bioc are included | ||
# CRAN | |||
# "https://cloud.r-project.org" | |||
# "https://bioconductor.org/packages/3.6/bioc" | |||
install.packages("biospear") # it will prompt to select CRAN | |||
install.packages("biospear", repos = "http://cran.rstudio.com") # NOT work since bioc repos is erased | |||
</pre> | |||
This will also install the '''BiocInstaller''' package if it has not been installed before. See also [https://www.bioconductor.org/install/ Install Bioconductor Packages]. | |||
=== Bioconductor packages depend on CRAN === | |||
For example [https://cran.r-project.org/web/packages/cowplot/index.html cowplot] shows ''breakpointR'' from Bioconductor depends on it. | |||
== update.packages() == | |||
update.packages(ask="graphics") can open a graphical window to select packages. | |||
== only upgrade to binary package versions == | |||
Try setting options(pkgType = "binary") before running update.packages(). To make this always be the default behavior, you can add that line to your .Rprofile file. | |||
How about options(install.packages.check.source = "no"), options(install.packages.compile.from.source = "never")? | |||
== Binary packages only for two versions of R == | |||
[https://www.r-bloggers.com/r-tip-check-what-repos-you-are-using/ Check What Repos You are Using]. | |||
'''CRAN only provides binaries for one version of R prior to the current one. So when CRAN moves to post-3.6.* R most non version-stuck mirrors will not have 3.5.* binary versions of packages.''' | |||
> | == Install a tar.gz (e.g. an archived package) from a local directory == | ||
<syntaxhighlight lang='bash'> | |||
R CMD INSTALL <package-name>.tar.gz | |||
> | </syntaxhighlight> | ||
Or in R: | |||
<syntaxhighlight lang='rsplus'> | |||
# Method 1: cannot install dependencies | |||
install.packages(<pathtopackage>, repos = NULL) | |||
# These paths can be source directories or archives or binary package | |||
# archive files (as created by ‘R CMD build --binary’). | |||
# (‘http://’ and ‘file://’ URLs are also accepted and the files | |||
# will be downloaded and installed from local copies.) | |||
# Method 2: take care of dependencies from CRAN | |||
devtools::install(<directory to package>, dependencies = TRUE) | |||
# this will use 'R CMD INSTALL' to install the package. | |||
# It will try to install dependencies of the package from CRAN, | |||
# if they're not already installed. | |||
</syntaxhighlight> | </syntaxhighlight> | ||
The installation process can be nasty due to the dependency issue. Consider the 'biospear' package | |||
<pre> | |||
> | biospear - plsRcox (archived) - plsRglm (archived) - bipartite | ||
- lars | |||
- pls | |||
- kernlab | |||
- mixOmics (CRAN->Bioconductor) | |||
- risksetROC | |||
- survcomp (Bioconductor) | |||
- rms | |||
</pre> | |||
So in order to install the 'plsRcox' package, we need to do the following steps. Note: plsRcox package is back on 6/2/2018. | |||
{{Pre}} | |||
# For curl | |||
system("apt update") | |||
" | system("apt install curl libcurl4-openssl-dev libssl-dev") | ||
# For X11 | |||
system("apt install libcgal-dev libglu1-mesa-dev libglu1-mesa-dev") | |||
system("apt install libfreetype6-dev") # https://stackoverflow.com/questions/31820865/error-in-installing-rgl-package | |||
</pre> | |||
# | |||
> | |||
{{Pre}} | |||
source("https://bioconductor.org/biocLite.R") | source("https://bioconductor.org/biocLite.R") | ||
biocLite(" | biocLite("survcomp") # this has to be run before the next command of installing a bunch of packages from CRAN | ||
# | |||
install.packages("https://cran.r-project.org/src/contrib/Archive/biospear/biospear_1.0.1.tar.gz", | |||
repos = NULL, type="source") | |||
# ERROR: dependencies ‘pkgconfig’, ‘cobs’, ‘corpcor’, ‘devtools’, ‘glmnet’, ‘grplasso’, ‘mboost’, ‘plsRcox’, | |||
# ‘pROC’, ‘PRROC’, ‘RCurl’, ‘survAUC’ are not available for package ‘biospear’ | |||
install.packages(c("pkgconfig", "cobs", "corpcor", "devtools", "glmnet", "grplasso", "mboost", | |||
== | "plsRcox", "pROC", "PRROC", "RCurl", "survAUC")) | ||
# optional: install.packages(c("doRNG", "mvnfast")) | |||
install.packages("https://cran.r-project.org/src/contrib/Archive/biospear/biospear_1.0.1.tar.gz", | |||
repos = NULL, type="source") | |||
# OR | |||
# devtools::install_github("cran/biospear") | |||
library(biospear) # verify | |||
</pre> | |||
To install the (deprecated, bioc) packages 'inSilicoMerging', | |||
{{Pre}} | |||
biocLite(c('rjson', 'Biobase', 'RCurl')) | |||
# destination directory is required | |||
# download.file("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoDb_2.7.0.tar.gz", | |||
. | # "~/Downloads/inSilicoDb_2.7.0.tar.gz") | ||
# download.file("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoMerging_1.15.0.tar.gz", | |||
# "~/Downloads/inSilicoMerging_1.15.0.tar.gz") | |||
# ~/Downloads or $HOME/Downloads won't work in untar() | |||
# untar("~/Downloads/inSilicoDb_2.7.0.tar.gz", exdir="/home/brb/Downloads") | |||
# untar("~/Downloads/inSilicoMerging_1.15.0.tar.gz", exdir="/home/brb/Downloads") | |||
# install.packages("~/Downloads/inSilicoDb", repos = NULL) | |||
# install.packages("~/Downloads/inSilicoMerging", repos = NULL) | |||
install.packages("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoDb_2.7.0.tar.gz", | |||
repos = NULL, type = "source") | |||
install.packages("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoMerging_1.15.0.tar.gz", | |||
repos = NULL, type = "source") | |||
</pre> | |||
=== R CMD INSTALL -l LIB/--library=LIB option === | |||
Install a package to a custom location | |||
<pre> | |||
$ R CMD INSTALL -l /usr/me/localR/library myRPackage.tar.gz | |||
</pre> | |||
Use a package installed in a custom location | |||
<pre> | <pre> | ||
R> library("myRPackage", lib.loc="/usr/me/local/R/library") | |||
# OR include below in .bashrc file | |||
$ export R_LIBS=/usr/me/local/R/library | |||
R> .libPaths() # check | |||
R> library("myRPackage") | |||
</pre> | |||
== Install a specific version of R/Bioconductor package == | |||
For packages from CRAN, use something like '''remotes::install_version'''("dplyr", "1.0.2") | |||
For packages from Bioconductor, see the two solutions [https://stackoverflow.com/questions/49487171/r-how-to-install-a-specified-version-of-a-bioconductor-package R how to install a specified version of a bioconductor package?] | |||
== Install multiple/different versions of the same R package == | |||
https://stackoverflow.com/a/2989369 | |||
<pre> | <pre> | ||
install.packages("~/Downloads/foo_0.1.1.tar.gz", lib = "/tmp", repos = NULL) | |||
# a new folder "/tmp/foo" will be created | |||
library(foo, lib.loc="/tmp") # Or use 'lib' to be consistent with install.packages() | |||
library(foo, lib.loc="~/dev/foo/v1") ## loads v1 | |||
# and | |||
library(foo, lib.loc="~/dev/foo/v2") ## loads v2 | |||
packageVersion("foo", lib.loc = "/tmp") | |||
sessionInfo() | |||
help(package = "foo", lib.loc = "/tmp") | |||
remove.packages("foo", lib = "/tmp") | |||
</pre> | </pre> | ||
The same works for install.packages(). help(install.packages) | |||
The install_version() from devtools and remotes will overwrite the existing installation. | |||
== Query an R package installed locally == | |||
<pre> | <pre> | ||
packageDescription("MASS") | |||
packageVersion("MASS") | |||
</pre> | </pre> | ||
== | == Query an R package (from CRAN) basic information: available.packages() == | ||
<syntaxhighlight lang='rsplus'> | |||
packageStatus() # Summarize information about installed packages | |||
available.packages() # List Available Packages at CRAN-like Repositories | |||
# Even I use an old version of R, it still return the latest version of the packages | |||
# The 'problem' happens on install.packages() too. | |||
</syntaxhighlight> | |||
The '''available.packages()''' command is useful for understanding package dependency. Use '''setRepositories()''' or 'RGUI -> Packages -> select repositories' to select repositories and '''options()$repos''' to check or change the repositories. | |||
<span style="color: red">The return result of available.packages() depends on the R version. </span> | |||
</ | |||
The | The number of packages returned from available.packages() is smaller than the number of packages obtained from '''rsync''' [https://cran.r-project.org/mirror-howto.html CRAN Mirror HOWTO/FAQ]. For example, 19473 < 25000. Some archived packages are still available via rsync. | ||
Also the '''packageStatus()''' is another useful function for query how many packages are in the repositories, how many have been installed, and individual package status (installed or not, needs to be upgraded or not). | |||
{{Pre}} | |||
> options()$repos | |||
CRAN | |||
"https://cran.rstudio.com/" | |||
> packageStatus() | |||
Number of installed packages: | |||
ok upgrade unavailable | |||
C:/Program Files/R/R-3.0.1/library 110 0 1 | |||
Number of available packages (each package counted only once): | |||
installed not installed | |||
... | http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0 76 4563 | ||
http://www.stats.ox.ac.uk/pub/RWin/bin/windows/contrib/3.0 0 5 | |||
http://www.bioconductor.org/packages/2.12/bioc/bin/windows/contrib/3.0 16 625 | |||
http://www.bioconductor.org/packages/2.12/data/annotation/bin/windows/contrib/3.0 4 686 | |||
> tmp <- available.packages() | |||
> str(tmp) | |||
... | chr [1:5975, 1:17] "A3" "ABCExtremes" "ABCp2" "ACCLMA" "ACD" "ACNE" "ADGofTest" "ADM3" "AER" ... | ||
- attr(*, "dimnames")=List of 2 | |||
..$ : chr [1:5975] "A3" "ABCExtremes" "ABCp2" "ACCLMA" ... | |||
..$ : chr [1:17] "Package" "Version" "Priority" "Depends" ... | |||
> tmp[1:3,] | |||
Package Version Priority Depends Imports LinkingTo Suggests | |||
A3 "A3" "0.9.2" NA "xtable, pbapply" NA NA "randomForest, e1071" | |||
ABCExtremes "ABCExtremes" "1.0" NA "SpatialExtremes, combinat" NA NA NA | |||
ABCp2 "ABCp2" "1.1" NA "MASS" NA NA NA | |||
Enhances License License_is_FOSS License_restricts_use OS_type Archs MD5sum NeedsCompilation File | |||
A3 NA "GPL (>= 2)" NA NA NA NA NA NA NA | |||
ABCExtremes NA "GPL-2" NA NA NA NA NA NA NA | |||
ABCp2 NA "GPL-2" NA NA NA NA NA NA NA | |||
Repository | |||
A3 "http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0" | |||
ABCExtremes "http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0" | |||
ABCp2 "http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0" | |||
</pre> | |||
And the following commands find which package depends on Rcpp and also which are from bioconductor repository. | |||
<syntaxhighlight lang='rsplus'> | |||
> pkgName <- "Rcpp" | |||
> rownames(tmp)[grep(pkgName, tmp[,"Depends"])] | |||
> tmp[grep("Rcpp", tmp[,"Depends"]), "Depends"] | |||
> ind <- intersect(grep(pkgName, tmp[,"Depends"]), grep("bioconductor", tmp[, "Repository"])) | |||
> rownames(grep)[ind] | |||
NULL | |||
> | > rownames(tmp)[ind] | ||
[1] "ddgraph" "DESeq2" "GeneNetworkBuilder" "GOSemSim" "GRENITS" | |||
[6] "mosaics" "mzR" "pcaMethods" "Rdisop" "Risa" | |||
[11] "rTANDEM" | |||
</syntaxhighlight> | </syntaxhighlight> | ||
== | == CRAN vs Bioconductor packages == | ||
<syntaxhighlight lang='rsplus'> | <syntaxhighlight lang='rsplus'> | ||
> R.version # 3.4.3 | |||
# CRAN | |||
> x <- available.packages() | |||
> dim(x) | |||
[1] 12581 17 | |||
# Bioconductor Soft | |||
< | > biocinstallRepos() | ||
BioCsoft | |||
"https://bioconductor.org/packages/3.6/bioc" | |||
BioCann | |||
"https://bioconductor.org/packages/3.6/data/annotation" | |||
BioCexp | |||
"https://bioconductor.org/packages/3.6/data/experiment" | |||
CRAN | |||
"https://cran.rstudio.com/" | |||
> y <- available.packages(repos = biocinstallRepos()[1]) | |||
> dim(y) | |||
[1] 1477 17 | |||
> intersect(x[, "Package"], y[, "Package"]) | |||
character(0) | |||
# Bioconductor Annotation | |||
> dim(available.packages(repos = biocinstallRepos()[2])) | |||
[1] 909 17 | |||
# Bioconductor Experiment | |||
> dim(available.packages(repos = biocinstallRepos()[3])) | |||
[1] 326 17 | |||
# CRAN + All Bioconductor | |||
> z <- available.packages(repos = biocinstallRepos()) | |||
> dim(z) | |||
[1] 15292 17 | |||
</syntaxhighlight> | |||
== | == Downloading Bioconductor package with an old R == | ||
When I try to download the [https://bioconductor.org/packages/release/bioc/html/GenomicDataCommons.html GenomicDataCommons] package using R 3.4.4 with Bioc 3.6 (the current R version is 3.5.0), it was found it can only install version 1.2.0 instead the latest version 1.4.1. | |||
== | It does not work by running biocLite("BiocUpgrade") to upgrade Bioc from 3.6 to 3.7. | ||
* http:// | <syntaxhighlight lang='rsplus'> | ||
* [http:// | source("https://bioconductor.org/biocLite.R") | ||
biocLite("BiocUpgrade") | |||
# Error: Bioconductor version 3.6 cannot be upgraded with R version 3.4.4 | |||
</syntaxhighlight> | |||
See some instruction on [https://packagemanager.rstudio.com/client/#/repos/3/overview RStudio package manager] website. | |||
== Analyzing data on CRAN packages == | |||
* New undocumented function in R 3.4.0: '''tools::CRAN_package_db()''' | |||
* http://blog.revolutionanalytics.com/2017/05/analyzing-data-on-cran-packages.html | |||
* [http://blog.schochastics.net/posts/2024-01-17_six-degrees-of-hadley-wickham/ Six degrees of Hadley Wickham: The CRAN co-authorship network] | |||
== R package location when they are installed by root == | |||
''/usr/local/lib/R/site-library'' | |||
== Customizing your package/library location == | |||
=== Add a personal directory to .libPaths() === | |||
<syntaxhighlight lang='rsplus'> | <syntaxhighlight lang='rsplus'> | ||
.libPaths( c( .libPaths(), "~/userLibrary") ) | |||
</syntaxhighlight> | </syntaxhighlight> | ||
No need to use the assignment operator. | |||
=== Install personal R packages after upgrade R, .libPaths(), Rprofile.site, R_LIBS_USER === | |||
{| class="wikitable" | |||
|- | |||
! File | |||
! Example | |||
|- | |||
| Rprofile.site/.Rprofile | |||
| .libPaths(c("/usr/lib/R/site-library",<br />       "/usr/lib/R/library")) | |||
|- | |||
| Renviron.site/.Renviron | |||
| R_LIB_SITE="/usr/lib/R/site-library:/usr/lib/R/library" | |||
|} | |||
Scenario: We already have installed many R packages under R 3.1.X in the user's directory. Now we upgrade R to a new version (3.2.X). We like to have these packages available in R 3.2.X. | |||
<span style="color:#0000FF">For Windows OS, refer to [http://cran.r-project.org/bin/windows/base/rw-FAQ.html#What_0027s-the-best-way-to-upgrade_003f R for Windows FAQ]</span> | |||
# | |||
</ | |||
The | The follow method works on Linux and Windows. | ||
<span style="color:#FF0000">Make sure only one instance of R is running</span> | |||
<pre> | |||
# Step 1. update R's built-in packages and install them on my personal directory | |||
update.packages(ask=FALSE, checkBuilt = TRUE, repos="http://cran.rstudio.com") | |||
# Step 2. update Bioconductor packages | |||
.libPaths() # The first one is my personal directory | |||
# [1] "/home/brb/R/x86_64-pc-linux-gnu-library/3.2" | |||
# [2] "/usr/local/lib/R/site-library" | |||
# [3] "/usr/lib/R/site-library" | |||
# [4] "/usr/lib/R/library" | |||
Sys.getenv("R_LIBS_USER") # may or may not equivalent to .libPaths()[1] | |||
ul <- unlist(strsplit(Sys.getenv("R_LIBS_USER"), "/")) | |||
src <- file.path(paste(ul[1:(length(ul)-1)], collapse="/"), "3.1") | |||
des <- file.path(paste(ul[1:(length(ul)-1)], collapse="/"), "3.2") | |||
pkg <- dir(src, full.names = TRUE) | |||
# | if (!file.exists(des)) dir.create(des) # If 3.2 subdirectory does not exist yet! | ||
file.copy(pkg, des, overwrite=FALSE, recursive = TRUE) | |||
source("http://www.bioconductor.org/biocLite.R") | |||
biocLite(ask = FALSE) | |||
</pre> | </pre> | ||
= | <span style="color:#0000FF">From Robert Kabacoff ([https://www.statmethods.net/interface/customizing.html R in Action])</span> | ||
* If you have a customized '''Rprofile.site file''' (see appendix B), save a copy outside of R. | |||
* Launch your current version of R and issue the following statements | |||
* | <pre> | ||
oldip <- installed.packages()[,1] | |||
* | save(oldip, file="path/installedPackages.Rdata") | ||
</pre> | |||
where ''path'' is a directory outside of R. | |||
* Download and install the newer version of R. | |||
* If you saved a customized version of the Rprofile.site file in step 1, copy it into the new installation. | |||
* Launch the new version of R, and issue the following statements | |||
<pre> | |||
load("path/installedPackages.Rdata") | |||
</ | newip <- installed.packages()[,1] | ||
for(i in setdiff(oldip, newip)) | |||
install.packages(i) | |||
</pre> | |||
where path is the location specified in step 2. | |||
* Delete the old installation (optional). | |||
This approach will install only packages that are available from the CRAN. It won’t find packages obtained from other locations. In fact, the process will display a list of packages that can’t be installed For example for packages obtained from Bioconductor, use the following method to update packages | |||
<pre> | |||
source(http://bioconductor.org/biocLite.R) | |||
biocLite(PKGNAME) | |||
</pre> | |||
== | === Persistent config and data for R packages with .Rprofile and .Renviron === | ||
[https:// | [https://blog.r-hub.io/2020/03/12/user-preferences/ Persistent config and data for R packages]. startup, rappdirs, hoardr, keyring. | ||
== | === Would you like to use a personal library instead? === | ||
Some posts from internet | |||
* | * [https://stackoverflow.com/questions/29969838/setting-r-libs-avoiding-would-you-like-to-use-a-personal-library-instead Setting R_LIBS & avoiding “Would you like to use a personal library instead?”]. Note: I try to create ~/.Renviron to add my personal folder in it. But update.packages() still asks me if I like to use a personal library instead (tested on Ubuntu + R 3.4). | ||
* [https://stackoverflow.com/questions/39879424/automatically-create-personal-library-in-r automatically create personal library in R]. Using suppressUpdates + specify '''lib''' in biocLite() or update.packages(Sys.getenv("R_LIBS_USER"), ask = F) <syntaxhighlight lang='rsplus'> | |||
# create local user library path (not present by default) | |||
dir.create(path = Sys.getenv("R_LIBS_USER"), showWarnings = FALSE, recursive = TRUE) | |||
# install to local user library path | |||
install.packages(p, lib = Sys.getenv("R_LIBS_USER"), repos = "https://cran.rstudio.com/") | |||
# Bioconductor version | |||
biocLite(p, suppressUpdates = TRUE, lib = Sys.getenv("R_LIBS_USER")) | |||
</syntaxhighlight> | |||
The problem can happen if the R was installed to the C:\Program Files\R folder by ''users'' but then some main packages want to be upgraded. R will always pops a message 'Would you like to use a personal library instead?'. | |||
To suppress the message and use the personal library always, | |||
[ | * Run R as administrator. If you do that, main packages can be upgraded from C:\Program Files\R\R-X.Y.Z\library folder. | ||
* [https:// | * [[Arraytools#Writable_R_package_directory_cannot_be_found|Writable R package directory cannot be found]] and a [[Arraytools#Download_required_R.2FBioconductor_.28software.29_packages|this]]. A solution here is to change the security of the R library folder so the user has a full control on the folder. | ||
* [https://cran.r-project.org/bin/windows/base/rw-FAQ.html#Does-R-run-under-Windows-Vista_003f Does R run under Windows Vista/7/8/Server 2008?] There are 3 ways to get around the issue. | |||
* [https:// | * [https://cran.r-project.org/bin/windows/base/rw-FAQ.html#I-don_0027t-have-permission-to-write-to-the-R_002d3_002e3_002e2_005clibrary-directory I don’t have permission to write to the R-3.3.2\library directory] | ||
library | |||
Actually the following hints will help us to create a convenient function UpdateMainLibrary() which will install updated main packages in the user's ''Documents'' directory without a warning dialog. | |||
* '''.libPaths()''' only returns 1 string "C:/Program Files/R/R-x.y.z/library" on the machines that does not have this problem | |||
* '''.libPaths()''' returns two strings "C:/Users/USERNAME/Documents/R/win-library/x.y" & "C:/Program Files/R/R-x.y.z/library" on machines with the problem. | |||
.. | |||
<syntaxhighlight lang='rsplus'> | <syntaxhighlight lang='rsplus'> | ||
UpdateMainLibrary <- function() { | |||
# Update main/site packages | |||
# The function is used to fix the problem 'Would you like to use a personal library instead?' | |||
> | if (length(.libPaths()) == 1) return() | ||
[ | |||
> install.packages( | ind_mloc <- grep("Program", .libPaths()) # main library e.g. 2 | ||
ind_ploc <- grep("Documents", .libPaths()) # personal library e.g. 1 | |||
if (length(ind_mloc) > 0L && length(ind_ploc) > 0L) | |||
# search outdated main packages | |||
old_mloc <- ! old.packages(.libPaths()[ind_mloc])[, "Package"] %in% | |||
installed.packages(.libPaths()[ind_ploc])[, "Package"] | |||
oldpac <- old.packages(.libPaths()[ind_mloc])[old_mloc, "Package"] | |||
if (length(oldpac) > 0L) | |||
install.packages(oldpac, .libPaths()[ind_ploc]) | |||
} | |||
</syntaxhighlight> | |||
On Linux, | |||
<syntaxhighlight lang='rsplus'> | |||
> update.packages() | |||
... | ... | ||
The downloaded source packages are in | The downloaded source packages are in | ||
‘/tmp/RtmpBrYccd/downloaded_packages’ | |||
Warning in install.packages(update[instlib == l, "Package"], l, contriburl = contriburl, : | |||
'lib = "/opt/R/3.5.0/lib/R/library"' is not writable | |||
Would you like to use a personal library instead? (yes/No/cancel) yes | |||
> | ... | ||
> system("ls -lt /home/brb/R/x86_64-pc-linux-gnu-library/3.5 | head") | |||
total 224 | |||
drwxrwxr-x 9 brb brb 4096 Oct 3 09:30 survival | |||
drwxrwxr-x 9 brb brb 4096 Oct 3 09:29 mgcv | |||
drwxrwxr-x 10 brb brb 4096 Oct 3 09:29 MASS | |||
drwxrwxr-x 9 brb brb 4096 Oct 3 09:29 foreign | |||
# So new versions of survival, mgc, MASS, foreign are installed in the personal directory | |||
# The update.packages() will issue warnings if we try to run it again. | |||
# It's OK to ignore these warnings. | |||
> update.packages() | |||
Warning: package 'foreign' in library '/opt/R/3.5.0/lib/R/library' will not be updated | |||
Warning: package 'MASS' in library '/opt/R/3.5.0/lib/R/library' will not be updated | |||
Warning: package 'mgcv' in library '/opt/R/3.5.0/lib/R/library' will not be updated | |||
Warning: package 'survival' in library '/opt/R/3.5.0/lib/R/library' will not be updated | |||
</syntaxhighlight> | |||
== | === R_LIBS_USER is empty in R 3.4.1 === | ||
See [[R#install.package.28.29_error.2C_R_LIBS_USER_is_empty_in_R_3.4.1|install.package() error, R_LIBS_USER is empty in R 3.4.1]]. | |||
== List vignettes from a package == | |||
< | <syntaxhighlight lang='rsplus'> | ||
vignette(package=PACKAGENAME) | |||
</syntaxhighlight> | |||
== List data from a package == | |||
<syntaxhighlight lang='rsplus'> | |||
data(package=PACKAGENAME) | |||
</syntaxhighlight> | |||
=== sysdata.rda === | |||
* [https://stackoverflow.com/a/24363504 Load data object when package is loaded] | |||
* https://r-pkgs.org/data.html | |||
== List all functions of a package == | |||
Assume a package is already loaded. Then | |||
<pre> | |||
ls("package:cowplot") | |||
</pre> | </pre> | ||
[http://www.cookbook-r.com/Scripts_and_functions/Getting_a_list_of_functions_and_objects_in_a_package/ Getting a list of functions and objects in a package]. This also assumes the package in loaded. In addition to functions (separated by primitive and non-primitive), it can show constants and objects. | |||
== List installed packages and versions == | |||
* http://heuristicandrew.blogspot.com/2015/06/list-of-user-installed-r-packages-and.html | |||
* [http://cran.r-project.org/web/packages/checkpoint/index.html checkpoint] package | |||
<syntaxhighlight lang='rsplus'> | |||
ip <- as.data.frame(installed.packages()[,c(1,3:4)]) | |||
rownames(ip) <- NULL | |||
unique(ip$Priority) | |||
# [1] <NA> base recommended | |||
# Levels: base recommended | |||
ip <- ip[is.na(ip$Priority),1:2,drop=FALSE] | |||
print(ip, row.names=FALSE) | |||
</syntaxhighlight> | |||
== Query the names of outdated packages == | |||
{{Pre}} | |||
psi <- packageStatus()$inst | |||
subset(psi, Status == "upgrade", drop = FALSE) | |||
# Package LibPath Version Priority Depends | |||
# RcppArmadillo RcppArmadillo C:/Users/brb/Documents/R/win-library/3.2 0.5.100.1.0 <NA> <NA> | |||
# Matrix Matrix C:/Program Files/R/R-3.2.0/library 1.2-0 recommended R (>= 2.15.2), methods | |||
# Imports LinkingTo Suggests | |||
# RcppArmadillo Rcpp (>= 0.11.0) Rcpp RUnit, Matrix, pkgKitten | |||
# Matrix graphics, grid, stats, utils, lattice <NA> expm, MASS | |||
# Enhances License License_is_FOSS License_restricts_use OS_type MD5sum | |||
# RcppArmadillo <NA> GPL (>= 2) <NA> <NA> <NA> <NA> | |||
# Matrix MatrixModels, graph, SparseM, sfsmisc GPL (>= 2) <NA> <NA> <NA> <NA> | |||
# NeedsCompilation Built Status | |||
# RcppArmadillo yes 3.2.0 upgrade | |||
# Matrix yes 3.2.0 upgrade | |||
</pre> | |||
== | The above output does not show the package version from the latest packages on CRAN. So the following snippet does that. | ||
[ | {{Pre}} | ||
psi <- packageStatus()$inst | |||
pl <- unname(psi$Package[psi$Status == "upgrade"]) # List package names | |||
ap <- as.data.frame(available.packages()[, c(1,2,3)], stringsAsFactors = FALSE) | |||
out <- cbind(subset(psi, Status == "upgrade")[, c("Package", "Version")], ap[match(pl, ap$Package), "Version"]) | |||
colnames(out)[2:3] <- c("OldVersion", "NewVersion") | |||
rownames(out) <- NULL | |||
out | |||
# Package OldVersion NewVersion | |||
# 1 RcppArmadillo 0.5.100.1.0 0.5.200.1.0 | |||
# 2 Matrix 1.2-0 1.2-1 | |||
</pre> | |||
To consider also the packages from Bioconductor, we have the following code. Note that "3.1" means the Bioconductor version and "3.2" is the R version. See [http://bioconductor.org/about/release-announcements/#release-versions Bioconductor release versions] page. | |||
{{Pre}} | {{Pre}} | ||
psic <- packageStatus(repos = c(contrib.url(getOption("repos")), | |||
"http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2", | |||
"http://www.bioconductor.org/packages/3.1/data/annotation/bin/windows/contrib/3.2"))$inst | |||
subset(psic, Status == "upgrade", drop = FALSE) | |||
pl <- unname(psic$Package[psic$Status == "upgrade"]) | |||
ap <- as.data.frame(available.packages(c(contrib.url(getOption("repos")), | |||
"http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2", | |||
"http://www.bioconductor.org/packages/3.1/data/annotation/bin/windows/contrib/3.2"))[, c(1:3)], | |||
stringAsFactors = FALSE) | |||
out <- cbind(subset(psic, Status == "upgrade")[, c("Package", "Version")], ap[match(pl, ap$Package), "Version"]) | |||
colnames(out)[2:3] <- c("OldVersion", "NewVersion") | |||
rownames(out) <- NULL | |||
out | |||
# Package OldVersion NewVersion | |||
# 1 limma 3.24.5 3.24.9 | |||
# 2 RcppArmadillo 0.5.100.1.0 0.5.200.1.0 | |||
# 3 Matrix 1.2-0 1.2-1 | |||
</pre> | </pre> | ||
== Searching for packages in CRAN == | |||
* [https://github.com/metacran/pkgsearch#readme pkgsearch] package - Search R packages on CRAN | |||
* | ** [https://rviews.rstudio.com/2019/03/01/some-r-packages-for-roc-curves/ Some R Packages for ROC Curves] | ||
* [http://blog.revolutionanalytics.com/2015/06/fishing-for-packages-in-cran.html Fishing for packages in CRAN] | |||
* [http://blog.revolutionanalytics.com/2017/01/cran-10000.html CRAN now has 10,000 R packages. Here's how to find the ones you need] | |||
* | * [https://rviews.rstudio.com/2018/10/22/searching-for-r-packages/ Searching for R packages], [https://cran.r-project.org/web/packages/packagefinder/index.html packagefinder] package <syntaxhighlight lang='rsplus'> | ||
library(packagefinder) | |||
findPackage("survival") | |||
# 272 out of 13256 CRAN packages found in 5 seconds | |||
* | findPackage("follic") | ||
# No results found. | |||
# Actually 'follic' comes from randomForestSRC package | |||
# https://www.rdocumentation.org/packages/randomForestSRC/versions/2.7.0 | |||
</syntaxhighlight> The result is shown in an html format with columns of SCORE, NAME, DESC_SHORT, DOWNL_TOTAL & GO. | |||
# | |||
== | == [http://www.r-pkg.org/ METACRAN (www.r-pkg.org)] - Search and browse all CRAN/R packages == | ||
* Source code on https://github.com/metacran. The 'PACKAGES' file is updated regularly to Github. | |||
* [https://stat.ethz.ch/pipermail/r-devel/2015-May/thread.html Announcement] on R/mailing list | |||
* | * Author's homepage on http://gaborcsardi.org/. | ||
== [https://cran.r-project.org/web/packages/cranly/ cranly] visualisations and summaries for R packages == | |||
[https://rviews.rstudio.com/2018/05/31/exploring-r-packages/ Exploring R packages with cranly] | |||
== Query top downloaded packages, download statistics == | |||
* Daily download statistics http://cran-logs.rstudio.com/. Note the page is split into 'package' download and 'R' download. It tracks | |||
** Package: date, time, size, r_version, r_arch, r_os, package, version, country, ip_id. | |||
** R: date, time, size, R version, os (win/src/osx), county, ip_id (reset daily). | |||
* Original methods | |||
** https://strengejacke.wordpress.com/2015/03/07/cran-download-statistics-of-any-packages-rstats/ | |||
** http://blog.revolutionanalytics.com/2015/06/working-with-the-rstudio-cran-logs.html | |||
* https://www.r-bloggers.com/finally-tracking-cran-packages-downloads/. The code still works. | |||
** [https://gist.github.com/arraytools/f037a4fd90ae8fe8e2896db6fe5b7fad My modified code] for showing the top download R packages. The original code suffers from memory issue (tested on my 64GB Linux box) when it is calling rbindlist() from data.table package with large data. Still 64GB is required since the matrix is 369M by 4 (12GB). | |||
** [http://www.bnosac.be/index.php/blog/12-popularity-bigdata-large-data-packages-in-r-and-ffbase-user-presentation Popularity bigdata / large data packages in R and ffbase useR presentation] | |||
* [https://blog.rsquaredacademy.com/introducing-pkginfo/ pkginfo]: Tools for Retrieving R Package Information. It's only in github. Shiny interface. | |||
* [https://www.r-pkg.org/downloaded Top 100 downloaded packages] from METACRAN | |||
=== cranlogs === | |||
[https://github.com/metacran/cranlogs cranlogs] package - Download Logs from the RStudio CRAN Mirror. Suitable on R console. | |||
* [https://colinfay.me/12-months-cranlogs/ 2018 through {cranlogs}] | |||
... | * [https://hadley.shinyapps.io/cran-downloads/ Shiny app] by Hadley (works for packages on CRAN only). It's like the Google Trend app. An example of collection: '''survC1, survAUC, TreatmentSelection, biospear , (glmnet)''' | ||
package | * [https://blog.r-hub.io/2019/05/02/cranlogs-2-1-1/ cranlogs 2.1.1 is on CRAN!] 5/2/2019 | ||
* [https://github.com/r-hub/cranlogs/issues/56 limit on number of packages as argument to cran_downloads] | |||
<pre> | |||
library(cranlogs) | |||
out <- cran_top_downloads("last-week", 100) # 100 is the maximum limit | |||
out$package | |||
</pre> | |||
=== packageRank === | |||
[https://github.com/lindbrook/packageRank packageRank] package: Computing and Visualizing CRAN Downloads. Suitable to run on RStudio cloud. Include both CRAN and Bioconductor. | |||
{{Pre}} | |||
> plot(cranDownloads(packages = c("packageRank", "limma"), when = "last-month")) | |||
> plot(cranDownloads(packages = c("shiny", "glmnet"), when = "last-month")) | |||
> plot(cranDownloads(packages = c("shiny", "glmnet"), from = "2019", to ="2019")) | |||
> plot(cranDownloads(packages = c("shiny", "glmnet"), from = "2019-12", to ="2019-12")) | |||
> plot(bioconductorDownloads(packages = c("edgeR", "DESeq2", "Rsubread", "limma"), when = "last-year")) | |||
</pre> | |||
=== BiocPkgTools === | |||
For Bioconductor packages, try [http://bioconductor.org/packages/release/bioc/html/BiocPkgTools.html BiocPkgTools]. See the [https://f1000research.com/articles/8-752 paper]. | |||
package | === dlstats === | ||
[https://cran.r-project.org/web/packages/dlstats/ dlstats]. Monthly download stats of 'CRAN' and 'Bioconductor' packages. | |||
=== Litmus: R Package Validation Reimagined === | |||
[https://www.r-bloggers.com/2025/03/should-i-use-your-r-package/ Should I Use Your R Package?] | |||
== installation path not writeable from running biocLite() == | |||
When I ran biocLite() to install a new package, I got a message (the Bioc packages are installed successfully anyway) | |||
<pre> | |||
... | |||
* DONE (curatedOvarianData) | |||
> | |||
package | The downloaded source packages are in | ||
‘/tmp/RtmpHxnH2K/downloaded_packages’ | |||
installation path not writeable, unable to update packages: rgl, rJava, | |||
codetools, foreign, lattice, MASS, spatial, survival | |||
</pre> | |||
However, if I uses install.package() it can update the package | |||
<syntaxhighlight lang='rsplus'> | |||
> packageVersion("survival") | |||
[1] ‘2.42.3’ | |||
> update.packages("survival") # Not working though no error message | |||
> packageVersion("survival") | |||
[1] ‘2.42.3’ | |||
> install.packages("survival") | |||
Installing package into ‘/home/brb/R/x86_64-pc-linux-gnu-library/3.4’ | |||
... | |||
* DONE (survival) | |||
The downloaded | The downloaded source packages are in | ||
‘/tmp/RtmpHxnH2K/downloaded_packages’ | |||
> | > packageVersion("survival") | ||
[1] ‘2.42.6’ | |||
> library(survival) | |||
> sessionInfo() # show survival package 2.42-6 was attached | |||
> library( | </syntaxhighlight> | ||
> | |||
It makes sense to always use personal directory when we install packages. See .libPaths(). | |||
== Warning: cannot remove prior installation of package == | |||
http://stackoverflow.com/questions/15932152/unloading-and-removing-a-loaded-package-withouth-restarting-r | |||
Instance 1. | |||
<pre> | |||
# Install the latest hgu133plus2cdf package | |||
# Remove/Uninstall hgu133plus2.db package | |||
# Put/Install an old version of IRanges (eg version 1.18.2 while currently it is version 1.18.3) | |||
# Test on R 3.0.1 | |||
library(hgu133plus2cdf) # hgu133pluscdf does not depend or import IRanges | |||
1 | source("http://bioconductor.org/biocLite.R") | ||
biocLite("hgu133plus2.db", ask=FALSE) # hgu133plus2.db imports IRanges | |||
# Warning:cannot remove prior installation of package 'IRanges' | |||
# Open Windows Explorer and check IRanges folder. Only see libs subfolder. | |||
</pre> | </pre> | ||
Note: | |||
* In the above example, all packages were installed under C:\Program Files\R\R-3.0.1\library\. | |||
* In another instance where I cannot reproduce the problem, new R packages were installed under C:\Users\xxx\Documents\R\win-library\3.0\. The different thing is IRanges package CAN be updated but if I use packageVersion("IRanges") command in R, it still shows the old version. | |||
* The above were tested on a desktop. | |||
Instance 2. | |||
{{Pre}} | |||
# On a fresh R 3.2.0, I install Bioconductor's depPkgTools & lumi packages. Then I close R, re-open it, | |||
# and install depPkgTools package again. | |||
> source("http://bioconductor.org/biocLite.R") | |||
> biocLite(" | Bioconductor version 3.1 (BiocInstaller 1.18.2), ?biocLite for help | ||
> biocLite("pkgDepTools") | |||
BioC_mirror: http://bioconductor.org | BioC_mirror: http://bioconductor.org | ||
Using Bioconductor version 3.1 (BiocInstaller 1.18. | Using Bioconductor version 3.1 (BiocInstaller 1.18.2), R version 3.2.0. | ||
Installing package(s) | Installing package(s) ‘pkgDepTools’ | ||
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/pkgDepTools_1.34.0.zip' | |||
Content type 'application/zip' length 390579 bytes (381 KB) | |||
downloaded 381 KB | |||
package ‘pkgDepTools’ successfully unpacked and MD5 sums checked | |||
Warning: cannot remove prior installation of package ‘pkgDepTools’ | |||
The downloaded binary packages are in | |||
C:\Users\brb\AppData\Local\Temp\RtmpYd2l7i\downloaded_packages | |||
> library(pkgDepTools) | |||
Error in library(pkgDepTools) : there is no package called ‘pkgDepTools’ | |||
</pre> | </pre> | ||
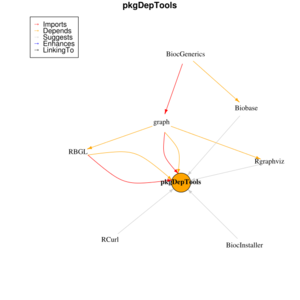
The pkgDepTools library folder appears in C:\Users\brb\Documents\R\win-library\3.2, but it is empty. The weird thing is if I try the above steps again, I cannot reproduce the problem. | |||
== Warning: dependency ‘XXX’ is not available == | |||
[https://stackoverflow.com/a/25721890 How should I deal with “package 'xxx' is not available (for R version x.y.z)” warning?] | |||
The | == Error: there is no package called XXX == | ||
The error happened when I try to run library() command on a package that was just installed. R 3.6.0. 'biospear' version is 1.0.2. macOS. | |||
{{Pre}} | |||
> library(biospear) | |||
Loading required package: pkgconfig | |||
Error: package or namespace load failed for ‘biospear’ in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vI[[i]]): | |||
there is no package called ‘mixOmics’ | |||
</pre> | </pre> | ||
Note | |||
== | * The package [https://cran.r-project.org/web/packages/mixOmics/index.html mixOmics] was removed from CRAN. It is now available on Bioconductor. | ||
* Tested to install on a docker container: '''docker run --net=host -it --rm r-base''' <syntaxhighlight lang='rsplus'> | |||
> install.packages(" | ERROR: dependency ‘car’ is not available for package ‘plsRglm’ | ||
* removing ‘/usr/local/lib/R/site-library/plsRglm’ | |||
ERROR: dependencies ‘plsRglm’, ‘mixOmics’, ‘survcomp’ are not available for package ‘plsRcox’ | |||
* removing ‘/usr/local/lib/R/site-library/plsRcox’ | |||
ERROR: dependencies ‘devtools’, ‘plsRcox’, ‘RCurl’ are not available for package ‘biospear’ | |||
* removing ‘/usr/local/lib/R/site-library/biospear’ | |||
</syntaxhighlight>The car package looks OK on CRAN. [https://cran.r-project.org/web/packages/survcomp/index.html survcomp] was moved from CRAN to Bioconductor too. | |||
* As we can see above, the official r-base image does not contain libraries enough to install RCurl/devtools packages. Consider the [https://hub.docker.com/r/rocker/tidyverse tidyverse] image (based on [https://github.com/rocker-org/rocker-versioned/blob/master/rstudio/Dockerfile RStudio image]) from the rocker project. <syntaxhighlight lang='bash'> | |||
docker pull rocker/tidyverse:3.6.0 | |||
docker run --net=host -it --rm -e PASSWORD=password -p 8787:8787 rocker/tidyverse:3.6.0 | |||
# the default username is 'rstudio' | |||
# Open a browser, log in. Run 'install.packages("RCurl")'. It works. | |||
</syntaxhighlight> | |||
* Testing on Mint linux also shows errors about dependencies of mixOmics and survcomp. | |||
* '''The best practice to install a package that may depend on packages located/moved to Bioconductor''': Run '''setRepositories(ind=1:2)''' before calling install.packages(). However, it does not '''remedy''' the situation that the 1st level imports package (eg plsRcox) was installed before but the 2nd level imports package (eg mixOmics) was not installed. | |||
* [https://stat.ethz.ch/R-manual/R-devel/library/tools/html/dependsOnPkgs.html dependsOnPkgs()]: Find Reverse Dependencies. It seems it only return packages that have been installed locally. For example, tools::dependsOnPkgs("RcppEigen", "LinkingTo") | |||
== Warning: Unable to move temporary installation == | |||
The problem seems to happen only on virtual machines (Virtualbox). | |||
* '''Warning: unable to move temporary installation `C:\Users\brb\Documents\R\win-library\3.0\fileed8270978f5\quadprog` to `C:\Users\brb\Documents\R\win-library\3.0\quadprog`''' when I try to run 'install.packages("forecast"). | |||
* '''Warning: unable to move temporary installation ‘C:\Users\brb\Documents\R\win-library\3.2\file5e0104b5b49\plyr’ to ‘C:\Users\brb\Documents\R\win-library\3.2\plyr’ ''' when I try to run 'biocLite("lumi")'. The other dependency packages look fine although I am not sure if any unknown problem can happen (it does, see below). | |||
Here is a note of my trouble shooting. | |||
# If I try to ignore the warning and load the lumi package. I will get an error. | |||
# If I try to run biocLite("lumi") again, it will only download & install lumi without checking missing 'plyr' package. Therefore, when I try to load the lumi package, it will give me an error again. | |||
# Even I install the plyr package manually, library(lumi) gives another error - missing mclust package. | |||
{{Pre}} | |||
> biocLite("lumi") | |||
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/BiocInstaller_1.18.2.zip' | |||
Content type 'application/zip' length 114097 bytes (111 KB) | |||
downloaded 111 KB | |||
... | |||
package ‘lumi’ successfully unpacked and MD5 sums checked | |||
The downloaded binary packages are in | |||
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages | |||
Old packages: 'BiocParallel', 'Biostrings', 'caret', 'DESeq2', 'gdata', 'GenomicFeatures', 'gplots', 'Hmisc', 'Rcpp', 'RcppArmadillo', 'rgl', | |||
'stringr' | |||
Update all/some/none? [a/s/n]: a | |||
also installing the dependencies ‘Rsamtools’, ‘GenomicAlignments’, ‘plyr’, ‘rtracklayer’, ‘gridExtra’, ‘stringi’, ‘magrittr’ | |||
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/Rsamtools_1.20.1.zip' | |||
Content type 'application/zip' length 8138197 bytes (7.8 MB) | |||
downloaded 7.8 MB | |||
Content type 'application/ | ... | ||
package ‘Rsamtools’ successfully unpacked and MD5 sums checked | |||
downloaded | package ‘GenomicAlignments’ successfully unpacked and MD5 sums checked | ||
package ‘plyr’ successfully unpacked and MD5 sums checked | |||
Warning: unable to move temporary installation ‘C:\Users\brb\Documents\R\win-library\3.2\file5e0104b5b49\plyr’ | |||
to ‘C:\Users\brb\Documents\R\win-library\3.2\plyr’ | |||
package ‘rtracklayer’ successfully unpacked and MD5 sums checked | |||
package ‘gridExtra’ successfully unpacked and MD5 sums checked | |||
package ‘stringi’ successfully unpacked and MD5 sums checked | |||
package ‘magrittr’ successfully unpacked and MD5 sums checked | |||
package ‘BiocParallel’ successfully unpacked and MD5 sums checked | |||
package ‘Biostrings’ successfully unpacked and MD5 sums checked | |||
Warning: cannot remove prior installation of package ‘Biostrings’ | |||
package ‘caret’ successfully unpacked and MD5 sums checked | |||
package ‘DESeq2’ successfully unpacked and MD5 sums checked | |||
package ‘gdata’ successfully unpacked and MD5 sums checked | |||
package ‘GenomicFeatures’ successfully unpacked and MD5 sums checked | |||
package ‘gplots’ successfully unpacked and MD5 sums checked | |||
package ‘Hmisc’ successfully unpacked and MD5 sums checked | |||
package ‘Rcpp’ successfully unpacked and MD5 sums checked | |||
package ‘RcppArmadillo’ successfully unpacked and MD5 sums checked | |||
package ‘rgl’ successfully unpacked and MD5 sums checked | |||
package ‘stringr’ successfully unpacked and MD5 sums checked | |||
> | The downloaded binary packages are in | ||
trying URL ' | C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages | ||
> library(lumi) | |||
Error in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vI[[i]]) : | |||
there is no package called ‘plyr’ | |||
Error: package or namespace load failed for ‘lumi’ | |||
> search() | |||
[1] ".GlobalEnv" "package:BiocInstaller" "package:Biobase" "package:BiocGenerics" "package:parallel" "package:stats" | |||
[7] "package:graphics" "package:grDevices" "package:utils" "package:datasets" "package:methods" "Autoloads" | |||
[13] "package:base" | |||
> biocLite("lumi") | |||
BioC_mirror: http://bioconductor.org | |||
Using Bioconductor version 3.1 (BiocInstaller 1.18.2), R version 3.2.0. | |||
Installing package(s) ‘lumi’ | |||
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/lumi_2.20.1.zip' | |||
Content type 'application/zip' length 18185326 bytes (17.3 MB) | |||
downloaded 17.3 MB | |||
package ‘lumi’ successfully unpacked and MD5 sums checked | |||
The downloaded binary packages are in | |||
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages | |||
> search() | > search() | ||
[1] ".GlobalEnv" | [1] ".GlobalEnv" "package:BiocInstaller" "package:Biobase" "package:BiocGenerics" "package:parallel" "package:stats" | ||
[7] "package:graphics" "package:grDevices" "package:utils" "package:datasets" "package:methods" "Autoloads" | |||
[7] | [13] "package:base" | ||
> library(lumi) | |||
[13] " | Error in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vI[[i]]) : | ||
there is no package called ‘plyr’ | |||
Error: package or namespace load failed for ‘lumi’ | |||
> biocLite("plyr") | |||
BioC_mirror: http://bioconductor.org | |||
Using Bioconductor version 3.1 (BiocInstaller 1.18.2), R version 3.2.0. | |||
Installing package(s) ‘plyr’ | |||
trying URL 'http://cran.rstudio.com/bin/windows/contrib/3.2/plyr_1.8.2.zip' | |||
Content type 'application/zip' length 1128621 bytes (1.1 MB) | |||
downloaded 1.1 MB | |||
package ‘plyr’ successfully unpacked and MD5 sums checked | |||
The downloaded binary packages are in | |||
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages | |||
= | > library(lumi) | ||
Error in loadNamespace(j <- i[[1L]], c(lib.loc, .libPaths()), versionCheck = vI[[j]]) : | |||
there is no package called ‘mclust’ | |||
Error: package or namespace load failed for ‘lumi’ | |||
> ?biocLite | |||
Warning messages: | |||
1: In read.dcf(file.path(p, "DESCRIPTION"), c("Package", "Version")) : | |||
cannot open compressed file 'C:/Users/brb/Documents/R/win-library/3.2/Biostrings/DESCRIPTION', probable reason 'No such file or directory' | |||
2: In find.package(if (is.null(package)) loadedNamespaces() else package, : | |||
there is no package called ‘Biostrings’ | |||
> library(lumi) | |||
Error in loadNamespace(j <- i[[1L]], c(lib.loc, .libPaths()), versionCheck = vI[[j]]) : | |||
there is no package called ‘mclust’ | |||
In addition: Warning messages: | |||
1: In read.dcf(file.path(p, "DESCRIPTION"), c("Package", "Version")) : | |||
cannot open compressed file 'C:/Users/brb/Documents/R/win-library/3.2/Biostrings/DESCRIPTION', probable reason 'No such file or directory' | |||
2: In find.package(if (is.null(package)) loadedNamespaces() else package, : | |||
there is no package called ‘Biostrings’ | |||
Error: package or namespace load failed for ‘lumi’ | |||
</pre> | |||
[http://r.789695.n4.nabble.com/unable-to-move-temporary-installation-td4521714.html Other people also have the similar problem]. The possible cause is the virus scanner locks the file and R cannot move them. | |||
Some possible solutions: | |||
#- | # Delete ALL folders under R/library (e.g. C:/Progra~1/R/R-3.2.0/library) folder and install the main package again using install.packages() or biocLite(). | ||
# | # For specific package like 'lumi' from Bioconductor, we can [[R#Bioconductor.27s_pkgDepTools_package|find out all dependency packages]] and then install them one by one. | ||
# | # Find out and install the top level package which misses dependency packages. | ||
## This is based on the fact that install.packages() or biocLite() '''sometimes''' just checks & installs the 'Depends' and 'Imports' packages and '''won't install all packages recursively''' | |||
# | ## we can do a small experiment by removing a package which is not directly dependent/imported by another package; e.g. 'iterators' is not dependent/imported by 'glment' directly but indirectly. So if we run '''remove.packages("iterators"); install.packages("glmnet")''', then the 'iterator' package is still missing. | ||
## A real example is if the missing packages are 'Biostrings', 'limma', 'mclust' (these are packages that 'minfi' directly depends/imports although they should be installed when I run biocLite("lumi") command), then I should just run the command '''remove.packages("minfi"); biocLite("minfi")'''. If we just run biocLite("lumi") or biocLite("methylumi"), the missing packages won't be installed. | |||
== Error in download.file(url, destfile, method, mode = "wb", ...) == | |||
HTTP status was '404 Not Found' | |||
Tested on an existing R-3.2.0 session. Note that VariantAnnotation 1.14.4 was just uploaded to Bioc. | |||
{{pre}} | |||
> biocLite("COSMIC.67") | |||
BioC_mirror: http://bioconductor.org | |||
Using Bioconductor version 3.1 (BiocInstaller 1.18.3), R version 3.2.0. | |||
Installing package(s) ‘COSMIC.67’ | |||
also installing the dependency ‘VariantAnnotation’ | |||
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/VariantAnnotation_1.14.3.zip' | |||
Error in download.file(url, destfile, method, mode = "wb", ...) : | |||
cannot open URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/VariantAnnotation_1.14.3.zip' | |||
In addition: Warning message: | |||
In download.file(url, destfile, method, mode = "wb", ...) : | |||
cannot open: HTTP status was '404 Not Found' | |||
Warning in download.packages(pkgs, destdir = tmpd, available = available, : | |||
download of package ‘VariantAnnotation’ failed | |||
installing the source package ‘COSMIC.67’ | |||
trying URL 'http://bioconductor.org/packages/3.1/data/experiment/src/contrib/COSMIC.67_1.4.0.tar.gz' | |||
Content type 'application/x-gzip' length 40999037 bytes (39.1 MB) | |||
</pre> | </pre> | ||
However, when I tested on a new R-3.2.0 (just installed in VM), the COSMIC package installation is successful. That VariantAnnotation version 1.14.4 was installed (this version was just updated today from Bioconductor). | |||
The cause of the error is the '''[https://github.com/wch/r-source/blob/trunk/src/library/utils/R/packages.R available.package()]''' function will read the rds file first from cache in a tempdir (C:\Users\XXXX\AppData\Local\Temp\RtmpYYYYYY). See lines 51-55 of <packages.R>. | |||
<pre> | |||
dest <- file.path(tempdir(), | |||
paste0("repos_", URLencode(repos, TRUE), ".rds")) | |||
if(file.exists(dest)) { | |||
res0 <- readRDS(dest) | |||
} else { | |||
... | |||
} | |||
</pre> | |||
Since my R was opened 1 week ago, the rds file it reads today contains old information. Note that Bioconductor does not hold the source code or binary code for the old version of packages. This explains why biocLite() function broke. When I restart R, the original problem is gone. | |||
If we look at the source code of available.packages(), we will see we could use '''cacheOK''' option in download.file() function. | |||
<pre> | |||
download.file(url, destfile, method, cacheOK = FALSE, quiet = TRUE, mode ="wb") | |||
</pre> | </pre> | ||
== | == Another case: Error in download.file(url, destfile, method, mode = "wb", ...) == | ||
{{Pre}} | |||
> install.packages("quantreg") | |||
There is a binary version available but the source version is later: | |||
binary source needs_compilation | |||
quantreg 5.33 5.34 TRUE | |||
Do you want to install from sources the package which needs compilation? | |||
y/n: n | |||
trying URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.4/quantreg_5.33.tgz' | |||
Warning in install.packages : | |||
cannot open URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.4/quantreg_5.33.tgz': HTTP status was '404 Not Found' | |||
Error in download.file(url, destfile, method, mode = "wb", ...) : | |||
cannot open URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.4/quantreg_5.33.tgz' | |||
Warning in install.packages : | |||
download of package ‘quantreg’ failed | |||
</pre> | |||
It seems the binary package cannot be found on the mirror. So the solution here is to download the package from the R main server. Note that after I have successfully installed the binary package from the main R server, I remove the package in R and try to install the binary package from rstudio.com server agin and it works this time. | |||
<pre> | |||
> install.packages("quantreg", repos = "https://cran.r-project.org") | |||
trying URL 'https://cran.r-project.org/bin/macosx/el-capitan/contrib/3.4/quantreg_5.34.tgz' | |||
Content type 'application/x-gzip' length 1863561 bytes (1.8 MB) | |||
================================================== | |||
downloaded 1.8 MB | |||
</pre> | |||
==== Another case: Error in download.file() on Windows 7 ==== | |||
For some reason, IE 8 cannot interpret https://ftp.ncbi.nlm.nih.gov though it understands ftp://ftp.ncbi.nlm.nih.gov. | |||
This is tested using R 3.4.3. | |||
{{Pre}} | |||
> download.file("https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", "test.soft.gz") | |||
trying URL 'https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz' | |||
Error in download.file("https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", : | |||
cannot open URL 'https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz' | |||
In addition: Warning message: | |||
In download.file("https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", : | |||
InternetOpenUrl failed: 'An error occurred in the secure channel support' | |||
> | > download.file("ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", "test.soft.gz") | ||
> | trying URL 'ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz' | ||
downloaded 9.1 MB | |||
</pre> | |||
== ERROR: failed to lock directory == | |||
/ | Follow the suggestion to remove the LOCK file. See the [https://stackoverflow.com/questions/23483303/error-failed-to-lock-directory-c-program-files-r-r-3-0-2-library-for-modifyi post]. | ||
It could happened in calling install.packages(), biocLite() or devtools::install_github(), and so on. | |||
== Error in unloadNamespace(package) == | |||
<pre> | <pre> | ||
> d3heatmap(mtcars, scale = "column", colors = "Blues") | |||
Error: 'col_numeric' is not an exported object from 'namespace:scales' | |||
> packageVersion("scales") | |||
[1] ‘0.2.5’ | |||
> library(scales) | |||
Error in unloadNamespace(package) : | |||
namespace ‘scales’ is imported by ‘ggplot2’ so cannot be unloaded | |||
In addition: Warning message: | |||
package ‘scales’ was built under R version 3.2.1 | |||
Error in library(scales) : | |||
library( | Package ‘scales’ version 0.2.4 cannot be unloaded | ||
> search() | |||
[1] ".GlobalEnv" "package:d3heatmap" "package:ggplot2" | |||
[4] "package:microbenchmark" "package:COSMIC.67" "package:BiocInstaller" | |||
[7] "package:stats" "package:graphics" "package:grDevices" | |||
[10] "package:utils" "package:datasets" "package:methods" | |||
[13] "Autoloads" "package:base" | |||
</pre> | </pre> | ||
If I open a new R session, the above error will not happen! | |||
The problem occurred because the 'scales' package version required by the d3heatmap package/function is old. See [https://github.com/rstudio/d3heatmap/issues/16 this post]. And when I upgraded the 'scales' package, it was ''locked'' by the package was ''imported'' by the ''ggplot2'' package. | |||
== | == Unload a package == | ||
Add '''unload = TRUE''' option to unload the namespace. See [https://www.rdocumentation.org/packages/base/versions/3.6.2/topics/detach detach()]. | |||
<pre> | |||
require(splines) | |||
''' | detach(package:splines, unload=TRUE) | ||
= | |||
</pre> | </pre> | ||
== | == crantastic == | ||
https:// | https://crantastic.org/. A community site for R packages where you can search for, review and tag CRAN packages. | ||
https://github.com/hadley/crantastic | |||
== New R packages as reported by [http://dirk.eddelbuettel.com/cranberries/ CRANberries] == | |||
http://blog.revolutionanalytics.com/2015/07/mranspackages-spotlight.html | |||
<pre> | <pre> | ||
# | #---------------------------- | ||
# SCRAPE CRANBERRIES FILES TO COUNT NEW PACKAGES AND PLOT | |||
# | |||
library(ggplot2) | |||
# Build a vextor of the directories of interest | |||
year <- c("2013","2014","2015") | |||
month <- c("01","02","03","04","05","06","07","08","09","10","11","12") | |||
span <-c(rep(month,2),month[1:7]) | |||
dir <- "http://dirk.eddelbuettel.com/cranberries" | |||
url2013 <- file.path(dir,"2013",month) | |||
< | url2014 <- file.path(dir,"2014",month) | ||
url2015 <- file.path(dir,"2015",month[1:7]) | |||
url <- c(url2013,url2014,url2015) | |||
< | |||
# Read each directory and count the new packages | |||
new_p <- vector() | |||
for(i in url){ | |||
raw.data <- readLines(i) | |||
new_p[i] <- length(grep("New package",raw.data,value=TRUE)) | |||
} | |||
# Plot | |||
time <- seq(as.Date("2013-01-01"), as.Date("2015-07-01"), by="months") | |||
new_pkgs <- data.frame(time,new_p) | |||
== | ggplot(new_pkgs, aes(time,y=new_p)) + | ||
geom_line() + xlab("") + ylab("Number of new packages") + | |||
geom_smooth(method='lm') + ggtitle("New R packages as reported by CRANberries") | |||
</pre> | |||
== R packages being removed == | |||
* | * https://dirk.eddelbuettel.com/cranberries/cran/removed/ | ||
* | * https://web.archive.org/ to find out the package website before it's removed | ||
== Top new packages in 2015 == | |||
* | * [http://opiateforthemass.es/articles/R-packages-in-2015/ 2015 R packages roundup] by CHRISTOPH SAFFERLING | ||
* [http://gforge.se/2016/01/r-trends-in-2015/ R trends in 2015] by MAX GORDON | |||
== keep.source.pkgs option == | |||
[https://blog.r-hub.io/2020/09/03/keep.source/ State of R packages in your library]. The original code formatting and commenting being removed by default, unless one changes some options for installing packages. | |||
* options(keep.source.pkgs = TRUE) | |||
* | * install.packages("rhub", INSTALL_opts = "--with-keep.source", type = "source") | ||
* R CMD install --with-keep.source | |||
[https://bugs.r-project.org/show_bug.cgi?id=18236 Package installation speed for packages installed with ‘keep.source’ has been improved.] 2021-12-2. | |||
== Speeding up package installation == | |||
# | * http://blog.jumpingrivers.com/posts/2017/speed_package_installation/ | ||
* [http://dirk.eddelbuettel.com/blog/2017/11/27/#011_faster_package_installation_one (Much) Faster Package (Re-)Installation via Caching] | |||
* [http://dirk.eddelbuettel.com/blog/2017/12/13/#013_faster_package_installation_two (Much) Faster Package (Re-)Installation via Caching, part 2] | |||
== An efficient way to install and load R packages == | |||
[https://www.statsandr.com/blog/an-efficient-way-to-install-and-load-r-packages/ An efficient way to install and load R packages] | |||
{{Pre}} | |||
# Package names | |||
packages <- c("ggplot2", "readxl", "dplyr", "tidyr", "ggfortify", "DT", "reshape2", "knitr", "lubridate", "pwr", "psy", "car", "doBy", "imputeMissings", "RcmdrMisc", "questionr", "vcd", "multcomp", "KappaGUI", "rcompanion", "FactoMineR", "factoextra", "corrplot", "ltm", "goeveg", "corrplot", "FSA", "MASS", "scales", "nlme", "psych", "ordinal", "lmtest", "ggpubr", "dslabs", "stringr", "assist", "ggstatsplot", "forcats", "styler", "remedy", "snakecaser", "addinslist", "esquisse", "here", "summarytools", "magrittr", "tidyverse", "funModeling", "pander", "cluster", "abind") | |||
# Install packages not yet installed | |||
installed_packages <- packages %in% rownames(installed.packages()) | |||
if (any(installed_packages == FALSE)) { | |||
install.packages(packages[!installed_packages]) | |||
} | |||
# Packages loading | |||
# | invisible(lapply(packages, library, character.only = TRUE)) | ||
</pre> | </pre> | ||
Alternatively use the '''pacman''' package. | |||
== library( , '''exclude, include.only''') == | |||
See [https://www.rdocumentation.org/packages/base/versions/3.6.2/topics/library ?library] | |||
<pre> | <pre> | ||
library(tidyverse) | |||
library(MASS, exclude='select') | |||
library(thepackage, include.only="thefunction") | |||
library( | |||
</pre> | </pre> | ||
== | == package ‘XXX’ was installed by an R version with different internals == | ||
it needs to be reinstalled for use with this R version. The problem seems to be specific to R 3.5.1 in Ubuntu 16.04. I got this message when I try to install the '''keras''' and '''tidyverse''' packages. The 'XXX' package includes ''nlme'' for installing "tidyverse" and ''Matrix'' for installing "reticulate". I have already logged in as root. I need to manually install these packages again though it seems I did not see a version change for these packages. | |||
Same error [https://stackoverflow.com/q/50992032 Error: package was installed by an R version with different internals; it needs to be reinstalled for use with this R version]. | |||
= R package dependencies = | Today it also happened when I tried to install "pec" which broke when it was installing "Hmisc". The error message is "Error : package ‘rpart’ was installed by an R version with different internals; it needs to be reinstalled for use with this R version". I am using R 3.5.2. rpart version is ‘4.1.13’. The solution is I install rpart again (under my account is enough) though rpart does not have a new version. Then I can install "Hmisc". | ||
= packrat and renv = | |||
See [[Reproducible#renv:_successor_to_the_packrat_package| Reproducible → packrat/renv]] | |||
= R package dependencies = | |||
== Depends, Imports, Suggests, Enhances, LinkingTo == | == Depends, Imports, Suggests, Enhances, LinkingTo == | ||
| Line 1,466: | Line 1,530: | ||
* Enhances: lists packages “enhanced” by the package at hand, e.g., by providing methods for classes from these packages, or ways to handle objects from these packages. | * Enhances: lists packages “enhanced” by the package at hand, e.g., by providing methods for classes from these packages, or ways to handle objects from these packages. | ||
* LinkingTo: A package that wishes to make use of '''header''' files in other packages needs to declare them as a comma-separated list in the field ‘LinkingTo’ in the DESCRIPTION file. | * LinkingTo: A package that wishes to make use of '''header''' files in other packages needs to declare them as a comma-separated list in the field ‘LinkingTo’ in the DESCRIPTION file. | ||
** An example is [https://www.bioconductor.org/packages/release/bioc/html/SingleR.html SingleR] that links to the ''beachmat'' package in its [https://github.com/LTLA/SingleR/blob/master/src/recompute_scores.cpp cpp source code]. | |||
== Package related functions from package 'utils' == | == Package related functions from package 'utils' == | ||
| Line 1,520: | Line 1,585: | ||
* [http://stat.ethz.ch/R-manual/R-devel/library/utils/html/chooseCRANmirror.html chooseCRANmirror()], [http://stat.ethz.ch/R-manual/R-devel/library/utils/html/chooseBioCmirror.html chooseBioCmirror()] | * [http://stat.ethz.ch/R-manual/R-devel/library/utils/html/chooseCRANmirror.html chooseCRANmirror()], [http://stat.ethz.ch/R-manual/R-devel/library/utils/html/chooseBioCmirror.html chooseBioCmirror()] | ||
* [http://stat.ethz.ch/R-manual/R-devel/library/utils/html/globalVariables.html suppressForeignCheck()] | * [http://stat.ethz.ch/R-manual/R-devel/library/utils/html/globalVariables.html suppressForeignCheck()] | ||
=== download.packages() for source package === | |||
Consider the [https://bioinformatics.mdanderson.org/estimate/rpackage.html esimate] package hosted on [https://r-forge.r-project.org/projects/estimate/ r-forge]. | |||
<pre> | |||
download.packages("estimate", destdir = "~/Downloads", | |||
repos = "https://R-Forge.R-project.org") | |||
</pre> | |||
== tools package == | == tools package == | ||
| Line 1,531: | Line 1,603: | ||
propRcpp <- nRcpp / nCompiled * 100 | propRcpp <- nRcpp / nCompiled * 100 | ||
</syntaxhighlight> | </syntaxhighlight> | ||
* package.dependencies(), pkgDepends(), etc are deprecated now, mostly in favor of package_dependencies() which is both more flexible and efficient. See [https://cran.rstudio.com/doc/manuals/r-release/NEWS.html R 3.3.0 News]. | * package.dependencies(), pkgDepends(), etc are deprecated now, mostly in favor of [https://www.rdocumentation.org/packages/tools/versions/3.6.2/topics/package_dependencies package_dependencies()] which is both more flexible and efficient. See [https://cran.rstudio.com/doc/manuals/r-release/NEWS.html R 3.3.0 News]. For example, tools::package_dependencies(c("remotes", "devtools"), recursive=TRUE) shows ''remotes'' has only a few dependencies while ''devtools'' has a lot. | ||
== crandep package == | |||
https://cran.r-project.org/web/packages/crandep/index.html. Useful to find reverse dependencies. [https://rdrr.io/cran/crandep/man/get_dep.html ?get_dep]. Consider the [https://cran.r-project.org/web/packages/abc/index.html abc] package: | |||
<pre> | |||
get_dep("abc", "depends") # abc depends on these packages | |||
# note my computer does not have 'abc' installed | |||
# from to type reverse | |||
# 1 abc abc.data depends FALSE | |||
# 2 abc nnet depends FALSE | |||
# 3 abc quantreg depends FALSE | |||
# 4 abc MASS depends FALSE | |||
# 5 abc locfit depends FALSE | |||
get_dep("abc", "reverse_depends") | |||
# from to type reverse | |||
# 1 abc abctools depends TRUE | |||
# 2 abc EasyABC depends TRUE | |||
x <- get_dep("RcppEigen", c("reverse linking to")) | |||
dim(x) | |||
# [1] 331 4 | |||
head(x, 3) | |||
# from to type reverse | |||
# 1 RcppEigen abess linking to TRUE | |||
# 2 RcppEigen acrt linking to TRUE | |||
# 3 RcppEigen ADMMnet linking to TRUE | |||
</pre> | |||
== How does this package depend on this other package == | |||
[https://www.r-bloggers.com/2023/08/three-four-r-functions-i-enjoyed-this-week/ How does this package depend on this other package?], [https://pak.r-lib.org/reference/pkg_deps_explain.html pak::pkg_deps_explain()] | |||
== pkgndep == | |||
* [https://cran.r-project.org/web/packages/pkgndep/index.html CRAN] & the paper [https://academic.oup.com/bioinformatics/advance-article/doi/10.1093/bioinformatics/btac449/6633919?login=false pkgndep: a tool for analyzing dependency heaviness of R packages] | |||
* [https://jokergoo.github.io/2023/11/16/package-dependencies-in-your-session/ Package dependencies in your session]. DESeq2 was used. | |||
== remotes == | |||
[https://www.rdocumentation.org/packages/remotes/versions/2.2.0/topics/package_deps remotes::local_package_deps(dependencies=TRUE)] will find and return all dependent packages based on the "DESCRIPTION" file. See an example [https://github.com/Bioconductor/OrchestratingSingleCellAnalysis here]. | |||
== Bioconductor's [http://www.bioconductor.org/packages/release/bioc/html/pkgDepTools.html pkgDepTools] package == | == Bioconductor's [http://www.bioconductor.org/packages/release/bioc/html/pkgDepTools.html pkgDepTools] package == | ||
| Line 1,748: | Line 1,857: | ||
</syntaxhighlight> | </syntaxhighlight> | ||
[[File:Genomicfeature dep dep.svg|300px]] [[File:Genomicalignments dep.svg|300px]] | [[File:Genomicfeature dep dep.svg|300px]] [[File:Genomicalignments dep.svg|300px]] | ||
=== Github repository === | |||
[https://thierrymoudiki.github.io/blog/2020/10/16/r/esgtoolkit-new-cran Submitting R package to CRAN] | |||
== cranlike == | == cranlike == | ||
| Line 1,754: | Line 1,866: | ||
cranlike keeps the package data in a SQLite database, in addition to the PACKAGES* files. This database is the canonical source of the package data. It can be updated quickly, to add and remove packages. The PACKAGES* files are generated from the database. | cranlike keeps the package data in a SQLite database, in addition to the PACKAGES* files. This database is the canonical source of the package data. It can be updated quickly, to add and remove packages. The PACKAGES* files are generated from the database. | ||
== [http://mran.revolutionanalytics.com/ MRAN] (CRAN only) == | == [http://mran.revolutionanalytics.com/ MRAN] (CRAN only) & checkpoint package == | ||
http://blog.revolutionanalytics.com/2014/10/explore-r-package-connections-at-mran.html | * http://blog.revolutionanalytics.com/2014/10/explore-r-package-connections-at-mran.html | ||
* [https://rsangole.netlify.app/post/2020/10/10/reproducible-work-in-r/?s=09 Reproducible Work in R] | |||
* [https://datacolada.org/108 MRAN is Dead, long live GRAN] 4/28/2023 | |||
According to the snapsot list [https://cran.microsoft.com/snapshot/ here], the oldest version is 2014-08-18 which corresponds to R 3.1.0. | According to the snapsot list [https://cran.microsoft.com/snapshot/ here], the oldest version is 2014-08-18 which corresponds to R 3.1.0. | ||
| Line 1,767: | Line 1,881: | ||
'''Note the Bioconductor packages have no similar solution.''' | '''Note the Bioconductor packages have no similar solution.''' | ||
== [https://cran.r-project.org/web/packages/ | == groundhog package == | ||
[https:// | * [https://cran.r-project.org/web/packages/groundhog/index.html groundhog]: Version-Control for CRAN, GitHub, and GitLab Packages (Bioconductor?) | ||
* It seems groundhog is like checkpoint but groundhog does not depend on MRAN and it will figure out the package dependencies by itself. | |||
* [https://www.brodrigues.co/blog/2023-01-12-repro_r/ MRAN is getting shutdown] - what else is there for reproducibility with R, or why reproducibility is on a continuum? The author provides an example where we can integrate "groundhog" in the Dockerfile for reproducibility. Pay attention to the sentence (appear 2 times) '''“why use Docker at all? Since it’s easy to install older versions of R on Windows and macOS, wouldn’t an renv.lock file suffice? Or even just {groundhog} which is arguably even easier to use?” ''' | |||
* [https://github.com/rocker-org/rocker-versioned2/issues/593 MRAN is getting shutdown #593] related to Rocker project. Rocker now uses [https://docs.posit.co/rspm/admin/ RSPM (PPM now)] from RStudio. | |||
* groundhog.library() differs from the library() function: 1) it installed and loaded packages in one step, 2) it allowed to install multiple packages. | |||
Note I use the docker's R since that's the easiest way to use an old version of R (e.g. for some old R script) in Linux. Note that it will create a new folder '''R_groundhog''' folder in the working directory (see the message below). | |||
<pre> | <pre> | ||
$ docker run --rm -it -v $(pwd):/home/docker \ | |||
-w /home/docker -u docker r-base:4.0.2 R | |||
> install.packages("groundhog") | |||
> library(groundhog") | |||
groundhog needs authorization to save files to '/home/docker/R_groundhog' | |||
Enter 'OK' to provide authorization | |||
OK | |||
The groundhog folder path is now: | |||
/home/docker/R_groundhog/groundhog_library/ | |||
Loaded 'groundhog' (version:2.1.0) using R-4.0.2 | |||
Tips and troubleshooting: https://groundhogR.com | |||
> groundhog.library(" | |||
library(ggplot2)", | |||
"2020-10-10") | |||
> library() | |||
# Packages in library ‘/usr/local/lib/R/site-library’: | |||
> packageVersion("ggplot2") | |||
[1] ‘3.3.2’ # the latest version is 3.4.0 on R 4.2.2 | |||
> library(ggplot2) | |||
</pre> | </pre> | ||
(Current groundhog is v3.1.0) If I don't specify a user in docker run, I'll need to call groundhog.library() twice in order to install packages (eg. "DT" package). In this case I can use '''groundhog:::save.cookie("copy_instead_of_renaming")''' before calling groundhog.library(). | |||
<pre> | |||
$ docker run -it --rm rocker/verse:4.3.0 bash | |||
root@58978695ec12:/# R | |||
R version 4.3.0 (2023-04-21) -- "Already Tomorrow" | |||
... | |||
> install.packages("groundhog"); library(groundhog) | |||
> .libPaths() | |||
[1] "/usr/local/lib/R/site-library" "/usr/local/lib/R/library" | |||
> get.groundhog.folder() | |||
[1] "/root/R_groundhog/groundhog_library/" | |||
== Install packages offline == | > groundhog:::save.cookie("copy_instead_of_renaming") | ||
http://www.mango-solutions.com/wp/2017/05/installing-packages-without-internet/ | groundhog needs authorization to save files to '/root/R_groundhog/' | ||
Enter 'OK' to provide authorization, and 'NO' not to. | |||
| >OK | |||
Groundhog folder set to: '/root/R_groundhog/groundhog_library/ | |||
--- You may change it with`set.groundhog.folder(<path>)`--- | |||
Downloading database with information for all CRAN packages ever published | |||
trying URL 'http://s3.wasabisys.com/groundhog/cran.toc.rds' | |||
Content type 'application/octet-stream' length 2010504 bytes (1.9 MB) | |||
================================================== | |||
downloaded 1.9 MB | |||
Downloading database with installation times for all source packages on CRAN | |||
trying URL 'http://s3.wasabisys.com/groundhog/cran.times.rds' | |||
Content type 'application/octet-stream' length 803912 bytes (785 KB) | |||
================================================== | |||
downloaded 785 KB | |||
> groundhog.library("DT", "2023-4-23") | |||
> find.package("DT") | |||
</pre> | |||
== rang == | |||
[http://blog.schochastics.net/post/rang-make-ancient-r-code-run-again/ rang: make ancient R code run again] | |||
== [https://cran.r-project.org/web/packages/cranly/ cranly] == | |||
[https://cran.r-project.org/web/packages/cranly/vignettes/dependence_trees.html R package dependence trees] | |||
== sessioninfo == | |||
<pre> | |||
tmp = session_info("sessioninfo") | |||
dim(tmp$packages) # [1] 7 11 | |||
tmp = session_info("tidyverse") | |||
dim(tmp$packages) # [1] 95 11 | |||
</pre> | |||
== Reverse dependence == | |||
* http://romainfrancois.blog.free.fr/index.php?post/2011/10/30/Rcpp-reverse-dependency-graph | |||
* [http://www.markvanderloo.eu/yaRb/2019/04/26/checking-reverse-dependencies-the-tiny-way/ Checking reverse dependencies: the tiny way] | |||
* [https://cran.r-project.org/web/packages/pkgcache/index.html pkgcache]: Cache 'CRAN'-Like Metadata and R Packages | |||
** [https://www.r-bloggers.com/2025/12/how-to-assess-usage-of-your-package/ How to Assess Usage of your Package] | |||
** We can calculate the number of direct, total and indirect dependencies | |||
== tinyverse == | |||
* https://www.tinyverse.org/ | |||
* [https://www.r-bloggers.com/2025/06/ggplot2-4-0-0-is-coming-and-why-ultimately-its-on-you-to-ensure-your-environments-are-reproducible/ ggplot2 4.0.0 is coming and why ultimately it’s on YOU to ensure your environments are reproducible] | |||
== Install packages offline == | |||
http://www.mango-solutions.com/wp/2017/05/installing-packages-without-internet/ | |||
== Install a packages locally and its dependencies == | == Install a packages locally and its dependencies == | ||
| Line 1,790: | Line 1,982: | ||
== A minimal R package (for testing purpose) == | == A minimal R package (for testing purpose) == | ||
https://github.com/joelnitta/minimal. Question: is there a one from CRAN? | * https://github.com/joelnitta/minimal. Question: is there a one from CRAN? | ||
* [https://kbroman.org/pkg_primer/pages/minimal.html The minimal R package] | |||
* Linux command '''ls -lSr | grep '^-' | head -5''' | |||
* [https://cran.r-project.org/web/packages/QuadRoot/ QuadRoot] - the smallest (7002 bytes) R package on CRAN as of 5/10/2023. Others include (starting from the smallest) relen, signibox, freqdist, fdq. | |||
* [https://github.com/cran/LifeInsuranceContracts LifeInsuranceContracts] - the smallest (1090 bytes). [https://github.com/cran/signibox signibox] - the 2nd smallest (1415 bytes) R package on CRAN as of 7/12/2025. | |||
== An R package that does not require others during install.packages() == | == An R package that does not require others during install.packages() == | ||
| Line 1,810: | Line 2,006: | ||
* [https://devguide.ropensci.org/ rOpenSci Packages: Development, Maintenance, and Peer Review] | * [https://devguide.ropensci.org/ rOpenSci Packages: Development, Maintenance, and Peer Review] | ||
* [https://b-rodrigues.github.io/modern_R/package-development.html Package development] from "Modern R with the tidyverse" | * [https://b-rodrigues.github.io/modern_R/package-development.html Package development] from "Modern R with the tidyverse" | ||
* [https://youtu.be/qxRSzDejea4 How to write your own R package] (video, 10 minutes) | |||
* [https://youtu.be/EpTkT6Rkgbs Building R packages with devtools and usethis | RStudio] (video, thomas mock) | |||
* [https://quantixed.org/2022/09/06/the-package-learning-how-to-build-an-r-package/ The Package: learning how to build an R package] | |||
* [https://www.mzes.uni-mannheim.de/socialsciencedatalab/article/r-package/ How to write your own R package and publish it on CRAN] | |||
* [https://www.r-bloggers.com/2024/08/r-package-development-workflow-assuming-youre-using-macos-or-linux/ R package development workflow (assuming you’re using macOS or Linux)] | |||
* [https://drmowinckels.io/blog/2025/rpackage-dev-calendar/ R Package Development Advent Calendar 2025: A Complete Journey] 2025 Dec. | |||
== Package structure == | == Package structure == | ||
http://r-pkgs.had.co.nz/package.html | http://r-pkgs.had.co.nz/package.html. On Linux/macOS, use '''tree -d DIRNAME''' to show the directories only. At a minimum, we will have '''R''' and '''man''' directories. | ||
* ChangeLog | * ChangeLog | ||
| Line 1,844: | Line 2,046: | ||
=== NAMESPACE and DESCRIPTION === | === NAMESPACE and DESCRIPTION === | ||
[https://stackoverflow.com/a/13261139 Namespace dependencies not required]. ''If you use import or importFrom in your NAMESPACE file, you should have an entry for that package in the Imports section of your DESCRIPTION file (unless there is a reason that you need to use Depends in which case the package should have an entry in Depends, and not Imports).'' | [https://stackoverflow.com/a/13261139 Namespace dependencies not required]. ''If you use import or importFrom in your NAMESPACE file, you should have an entry for that package in the Imports section of your DESCRIPTION file (unless there is a reason that you need to use Depends in which case the package should have an entry in Depends, and not Imports).'' | ||
=== license === | |||
* [https://stackoverflow.com/a/38549043 What kind of license is the best license for an R package?] | |||
* https://cran.r-project.org/web/licenses/ | |||
* https://r-pkgs.org/description.html#description-license | |||
* [https://win-vector.com/2019/07/30/some-notes-on-gnu-licenses-in-r-packages/ Some Notes on GNU Licenses in R Packages] | |||
== Install software for PDF output == | == Install software for PDF output == | ||
| Line 1,851: | Line 2,059: | ||
== Windows: Rtools == | == Windows: Rtools == | ||
[https://thecoatlessprofessor.com/programming/installing-rtools-for-compiled-code-via-rcpp/ Installing RTools for Compiled Code via Rcpp]. Just remember to check the option to include some paths in the '''PATH''' environment variable. | <ul> | ||
<li>(Old) [https://thecoatlessprofessor.com/programming/installing-rtools-for-compiled-code-via-rcpp/ Installing RTools for Compiled Code via Rcpp]. Just remember to check the option to include some paths in the '''PATH''' environment variable. | |||
<li>[https://cran.rstudio.com/bin/windows/Rtools/rtools40.html Using Rtools4 on Windows]. This version gives more details about the story in Rtools4X versions. | |||
* https://cloud.r-project.org/bin/windows/base/howto-R-4.3.html | |||
<li>Testing Rtools43 on R 4.3.3. According to the information on the website: '''When using R installed by the installer, no further setup is necessary after installing Rtools43 to build R packages from source.''' I can testify that by running ''install.packages("jsonlite", type = "source")'' . We can also verify that make can be found. | |||
<pre> | |||
Sys.which("make") | |||
## "C:\\rtools40\\usr\\bin\\make.exe" | |||
</pre> | |||
</ul> | |||
Screenshots of installation of Rtools44 | |||
[[File:Rtools44.png|350px]] | |||
== R CMD == | == R CMD == | ||
[https://cran.r-project.org/doc/manuals/R-admin.html R Installation and Administration] | [https://cran.r-project.org/doc/manuals/R-admin.html R Installation and Administration] | ||
| Line 1,867: | Line 2,087: | ||
* https://usethis.r-lib.org/. '''usethis''' is a workflow package: it automates repetitive tasks that arise during project setup and development, both for R packages and non-package projects. | * https://usethis.r-lib.org/. '''usethis''' is a workflow package: it automates repetitive tasks that arise during project setup and development, both for R packages and non-package projects. | ||
** [https://youtu.be/79s3z0gIuFU?t=705 R Package Development 1: Where to Start] by John Muschelli. [https://www.youtube.com/playlist?list=PLk3B5c8iCV-T4LM0mwEyWIunIunLyEjqM R Package Development playlist]. | ** [https://youtu.be/79s3z0gIuFU?t=705 R Package Development 1: Where to Start] by John Muschelli. [https://www.youtube.com/playlist?list=PLk3B5c8iCV-T4LM0mwEyWIunIunLyEjqM R Package Development playlist]. | ||
** [https://usethis.r-lib.org/reference/create_package.html create_package()] | ** [https://usethis.r-lib.org/reference/create_package.html create_package(path)] with the package name of your choice. | ||
** [https://javirudolph.github.io/RLadies-Gainesville-FL/20190429-Rudolph-packages/apr29presentation.html#1 Sharing with others and helping yourself] (slides, use right click menu to save as an HTML file) | |||
* [https://www.tidyverse.org/articles/2019/04/usethis-1.5.0/ usethis 1.5.0] | * [https://www.tidyverse.org/articles/2019/04/usethis-1.5.0/ usethis 1.5.0] | ||
* [https://www.tidyverse.org/blog/2020/04/usethis-1-6-0/ usethis 1.6.0] | * [https://www.tidyverse.org/blog/2020/04/usethis-1-6-0/ usethis 1.6.0] | ||
* [https://www.tidyverse.org/blog/2020/12/usethis-2-0-0/ usethis 2.0.0] | |||
=== Github Actions === | === Github Actions === | ||
| Line 1,877: | Line 2,099: | ||
* Then I further delete the 'devel' line (line 23) to reduce one more platform to check. This took 3m to check. My example is on [https://github.com/arraytools/rtoy/actions Github] (rtoy). | * Then I further delete the 'devel' line (line 23) to reduce one more platform to check. This took 3m to check. My example is on [https://github.com/arraytools/rtoy/actions Github] (rtoy). | ||
I also try to follow [https://ropenscilabs.github.io/actions_sandbox/websites-using-pkgdown-bookdown-and-blogdown.html Github Actions with R] and create a [https://github.com/r-lib/pkgdown pkgdown]/package documentation web page. | I also try to follow [https://ropenscilabs.github.io/actions_sandbox/websites-using-pkgdown-bookdown-and-blogdown.html Github Actions with R] and create a [https://github.com/r-lib/pkgdown pkgdown]/package documentation web page. | ||
* '''usethis::use_github_actions("pkgdown")''' Now go to Github repo's Settings -> Options and scroll down until you see ''Github Pages''. For ''Source'', the page site should be set to being built from the root folder of the ''gh-pages''. | |||
* I have used Jekyll to create a gh-page. I don't need to delete anything for this new gh-pages. I just need to go to the repository setting and (scroll down until we see Github Pages) change the Source of Github Pages to 'gh-pages branch' from 'master branch'. | * I have used Jekyll to create a gh-page. I don't need to delete anything for this new gh-pages. I just need to go to the repository setting and (scroll down until we see Github Pages) change the Source of Github Pages to 'gh-pages branch' from 'master branch'. | ||
* The files on the gh-pages branch are generated by Github Actions; these files are not available on my local machine. My location machine only has '''.github/workflows/pkgdown.yaml''' file. | * The files on the gh-pages branch are generated by Github Actions; these files are not available on my local machine. My location machine only has '''.github/workflows/pkgdown.yaml''' file. | ||
| Line 1,884: | Line 2,107: | ||
* The workflow file specifies R version and OS platform. | * The workflow file specifies R version and OS platform. | ||
* Right now the workflow file (like pkgdown) is using "r-lib/actions/setup-r@master" that has an "action.yml" file. The r-version is '3.x' only. What about if R 4.0.0 is released? | * Right now the workflow file (like pkgdown) is using "r-lib/actions/setup-r@master" that has an "action.yml" file. The r-version is '3.x' only. What about if R 4.0.0 is released? | ||
=== Packages, webpages and Github === | |||
[https://debruine.github.io/tutorials/index.html Tutorials] by Lisa DeBruine | |||
== biocthis == | |||
[https://lcolladotor.github.io/biocthis/ biocthis], [https://speakerdeck.com/lcolladotor/biocthis-tab slides] | |||
== R package depends vs imports == | == R package depends vs imports == | ||
| Line 1,930: | Line 2,159: | ||
</syntaxhighlight> | </syntaxhighlight> | ||
== Useful functions for accessing files in packages | == Useful functions for accessing files in packages == | ||
* [https://stat.ethz.ch/R-manual/R-devel/library/base/html/system.file.html system.file()] | * [https://stat.ethz.ch/R-manual/R-devel/library/base/html/system.file.html system.file()] | ||
* [https://stat.ethz.ch/R-manual/R-devel/library/base/html/find.package.html path.package()] | * [https://stat.ethz.ch/R-manual/R-devel/library/base/html/find.package.html path.package() & find.package()]. Note that path.package() requires the package to be loaded. | ||
* [https://www.rdocumentation.org/packages/base/versions/3.6.2/topics/file.path file.path()]. This is not related to packages. | |||
* [https://www.rdocumentation.org/packages/base/versions/3.6.2/topics/normalizePath normalizePath()]. This is not related to packages. | |||
{{Pre}} | {{Pre}} | ||
> system.file(package = "batr") | > system.file(package = "batr") | ||
[1] "f:/batr" | [1] "f:/batr" | ||
> system.file("extdata", package = " | > system.file("extdata", "logo.png", package = "cowplot") # Mac | ||
[1] "/Library/Frameworks/R.framework/Versions/4.0/Resources/library/cowplot/extdata/logo.png" | |||
> path.package("batr") | > path.package("batr") | ||
[1] "f:\\batr" | [1] "f:\\batr" | ||
> path.package("ggplot2") # Mac | |||
[1] "/Library/Frameworks/R.framework/Versions/4.0/Resources/library/ggplot2" | |||
# sometimes it returns the forward slash format for some reason; C:/Program Files/R/R-3.4.0/library/batr | # sometimes it returns the forward slash format for some reason; C:/Program Files/R/R-3.4.0/library/batr | ||
# so it is best to add normalizePath(). | # so it is best to add normalizePath(). | ||
> normalizePath(path.package("batr")) | > normalizePath(path.package("batr")) | ||
> file.path("f:", "git", "surveyor") | |||
[1] "f:/git/surveyor" | |||
</pre> | </pre> | ||
== Internal functions == | == Internal functions == | ||
* [https://blog.r-hub.io/2019/12/12/internal-functions/ Internal functions in R packages] | * [https://blog.r-hub.io/2019/12/12/internal-functions/ Internal functions in R packages] | ||
* [https://statisticaloddsandends.wordpress.com/2021/06/10/documentation-for-internal-functions/ Documentation for internal functions] | |||
== RStudio shortcuts == | == RStudio shortcuts == | ||
| Line 1,955: | Line 2,193: | ||
[https://cran.r-project.org/web/packages/available/index.html available]. Check if a given package name is available to use. It checks the name's validity. Checks if it is used on 'GitHub', 'CRAN' and 'Bioconductor'. | [https://cran.r-project.org/web/packages/available/index.html available]. Check if a given package name is available to use. It checks the name's validity. Checks if it is used on 'GitHub', 'CRAN' and 'Bioconductor'. | ||
== Create R package with usethis == | == Create an R package == | ||
* [https://www.pipinghotdata.com/posts/2020-10-25-your-first-r-package-in-1-hour/?s=09 Your first R package in 1 hour] | |||
* [https://www.r-bloggers.com/2025/03/learning-to-create-an-r-package-with-deliberate-redundancy-%f0%9f%a4%a3-a-note-for-myself/ Learning To Create an R Package With Deliberate Redundancy - A Note For Myself] | |||
=== Using usethis === | |||
[https://trinostics.blogspot.com/2020/04/new-r-package-foo-updated.html New R Package 'foo' -- Updated] | [https://trinostics.blogspot.com/2020/04/new-r-package-foo-updated.html New R Package 'foo' -- Updated] | ||
# setwd() | # setwd() | ||
# usethis::create_package("foo") | # usethis::create_package("foo") | ||
# usethis::use_git(); usethis_github() | |||
# usethis::use_mit_license("Your name") | |||
# usethis::use_r("foo_function") | |||
# usethis::use_package("dplyr") # specify import dependency, e.g. dplyr package | |||
# usethis::use_testthat() | # usethis::use_testthat() | ||
# usethis::use_test("firsttest") | # usethis::use_test("firsttest") | ||
| Line 1,964: | Line 2,210: | ||
# roxygen2::roxygenise() | # roxygen2::roxygenise() | ||
== | === Using [https://github.com/hadley/devtools devtools] and usethis === | ||
* https://www.r-project.org/nosvn/pandoc/devtools.html | * https://www.r-project.org/nosvn/pandoc/devtools.html | ||
| Line 1,972: | Line 2,218: | ||
* [https://blog.r-hub.io/2020/04/29/maintenance/ Workflow automation tools for package developers] | * [https://blog.r-hub.io/2020/04/29/maintenance/ Workflow automation tools for package developers] | ||
* [https://jozefhajnala.gitlab.io/r/r102-addin-roxytags/ RStudio:addins part 2 - roxygen documentation formatting made easy] | * [https://jozefhajnala.gitlab.io/r/r102-addin-roxytags/ RStudio:addins part 2 - roxygen documentation formatting made easy] | ||
** Inserting a skeleton - Do this by placing your cursor anywhere in the function you want to document and click Code Tools -> Insert Roxygen Skeleton | |||
* [http://web.mit.edu/insong/www/pdf/rpackage_instructions.pdf Instructions for Creating Your Own R Package]. It includes creating a R package with functions written in C++ via Rcpp helper function. | * [http://web.mit.edu/insong/www/pdf/rpackage_instructions.pdf Instructions for Creating Your Own R Package]. It includes creating a R package with functions written in C++ via Rcpp helper function. | ||
* [https://www.r-bloggers.com/2025/09/creating-a-simple-r-package-with-c-code-to-sum-x-1/ Creating a simple R package with C++ code to sum “x + 1”] by '''usethis''' and '''devtools'''. | |||
* [https://www.rstudio.com/wp-content/uploads/2015/06/devtools-cheatsheet.pdf devtools cheatsheet] (2 pages) | * [https://www.rstudio.com/wp-content/uploads/2015/06/devtools-cheatsheet.pdf devtools cheatsheet] (2 pages) | ||
* My first R package [https://qualityandinnovation.com/2019/10/13/my-first-r-package-part-1/ Part 1], [https://qualityandinnovation.com/2019/10/13/my-first-r-package-part-2/ Part 2], [https://qualityandinnovation.com/2019/10/13/my-first-r-package-part-3/ Part 3]. It uses 3 packages: usethis, roxygen2 and devtools. | * My first R package [https://qualityandinnovation.com/2019/10/13/my-first-r-package-part-1/ Part 1], [https://qualityandinnovation.com/2019/10/13/my-first-r-package-part-2/ Part 2], [https://qualityandinnovation.com/2019/10/13/my-first-r-package-part-3/ Part 3]. It uses 3 packages: usethis, roxygen2 and devtools. | ||
* [https://twitter.com/JennyBryan/status/1364288709265215489?s=20 If you love devtools::load_all(), pkgload is where the magic actually happens]. [https://r-lib.github.io/pkgload/news/index.html pkgload] package. | |||
A Typical Streamlined Workflow | |||
<ol> | |||
<li>Start a New Package | |||
* Navigate to the directory where you want your package to live. | |||
* Run the following command in your R console: <code><nowiki>usethis::create_package("my_package")</nowiki></code> | |||
* This creates the '''my_package''' directory with the basic file structure and a '''.Rproj''' file. | |||
<li>Open the Project | |||
* Open the newly created '''my_package.Rproj''' file in RStudio. This is the most crucial step as it sets your working directory to the package root. | |||
<li>Add Your First R Script | |||
* Use this command to create a new R script file: <code><nowiki>usethis::use_r("my_function")</nowiki></code> | |||
* This creates the '''R/my_function.R''' file and opens it for you. | |||
<li>Write and Document Your Code | |||
* Add your function code to '''R/my_function.R'''. | |||
* Use roxygen2 comments to document your function. | |||
<li>Add a Test File | |||
* Use this command to create a basic test file for your function: <code><nowiki>usethis::use_test("my_function")</nowiki></code> | |||
* This creates a '''test-my_function.R''' file in the '''tests/testthat/''' directory. | |||
<li>Interactive Development Loop | |||
* Use these commands repeatedly as you work: <code><nowiki>devtools::load_all() | |||
devtools::test()</nowiki></code> | |||
<li>Generate Documentation | |||
* After writing your roxygen comments, run this command to generate the documentation: <code><nowiki>devtools::document()</nowiki></code> | |||
<li>Setup the README | |||
* Run this command to create a <code>README.Rmd</code> file: <code><nowiki>usethis::use_readme_rmd()</nowiki></code> | |||
<li>Write the README Content | |||
* Open the <code>README.Rmd</code> file and add your package description, installation instructions, and examples. | |||
<li>Build the README | |||
* After making changes to the '''.Rmd''' file, run this command to generate the '''README.md''' file: <code><nowiki>devtools::build_readme()</nowiki></code> | |||
<li>Perform a Final Check | |||
* Before sharing, run a comprehensive check: <code><nowiki>devtools::check()</nowiki></code> | |||
* This will report any issues that need to be fixed. | |||
<li>Version Control and Sharing | |||
* To use Git and GitHub: <code><nowiki>usethis::use_git() | |||
</ | usethis::use_github()</nowiki></code> | ||
* The rendered '''README.md''' will be displayed on your repository's front page. | |||
<li>Release | |||
* For a release, use one of these commands: <code><nowiki>devtools::install_github("your_username/my_package") # share with others | |||
# | devtools::release() # submitting to CRAN</nowiki></code> | ||
</ol> | |||
== | === Using RStudio === | ||
<ul> | <ul> | ||
<li>[https://support.rstudio.com/hc/en-us/articles/200486508-Building-Testing-and-Distributing-Packages Building, Testing, and Distributing Packages] from RStudio. <BR> | <li>[https://support.rstudio.com/hc/en-us/articles/200486508-Building-Testing-and-Distributing-Packages Building, Testing, and Distributing Packages] from RStudio. <BR> | ||
| Line 2,034: | Line 2,293: | ||
** How to publish your package to Github | ** How to publish your package to Github | ||
* [https://technistema.com/posts/building-a-corporate-r-package-for-pleasure-and-profit/ Building a Corporate R Package for Pleasure and Profit] | * [https://technistema.com/posts/building-a-corporate-r-package-for-pleasure-and-profit/ Building a Corporate R Package for Pleasure and Profit] | ||
* Toy example | * Toy example* | ||
*# In RStudio, click "File" - "New Project" - "New Directory" - "R package". Package name = "a1" (this naming will guarantee this package will be shown on top of all R packages in RStudio). Press 'Create Project'. This new project folder will have necessary files for an R package including DESCRIPTION, NAMESPACE, R/hello.R, man/hello.Rd. The "hello.R" file will be opened in RStudio automatically. | *# In RStudio, click "File" - "New Project" - "New Directory" - "R package". Package name = "a1" (this naming will guarantee this package will be shown on top of all R packages in RStudio). Press 'Create Project'. This new project folder will have necessary files for an R package including DESCRIPTION, NAMESPACE, R/hello.R, man/hello.Rd. The "hello.R" file will be opened in RStudio automatically. | ||
*# Create a new R file under a1/R folder. An example of this R file <add.R> containing roxygen comments can be found under [http://r-pkgs.had.co.nz/man.html here]. Pressing Ctrl/Cmd + Shift + D (or running '''devtools::document()''') will generate a man/add.Rd. | *# Create a new R file under a1/R folder. An example of this R file <add.R> containing roxygen comments can be found under [http://r-pkgs.had.co.nz/man.html here]. Pressing Ctrl/Cmd + Shift + D (or running '''devtools::document()''') will generate a man/add.Rd. | ||
| Line 2,042: | Line 2,301: | ||
*# In RStudio, type '''help(package = "a1") ''' or click "Packages" tab on the bottom-right panel and click "a1" package. It will show a line "User guides, package vignettes and other documentation". The vignette we just created will be available in HTML, source and R code format. | *# In RStudio, type '''help(package = "a1") ''' or click "Packages" tab on the bottom-right panel and click "a1" package. It will show a line "User guides, package vignettes and other documentation". The vignette we just created will be available in HTML, source and R code format. | ||
== | === Using RStudio.cloud === | ||
There is a problem. We can use devtools::create("/cloud/project") to create a new package. When it builds the source package, the package file will be located in the root directory of the package. However, if we use a local RStudio to create a source package, the source package will be located in the upper directory. | There is a problem. We can use devtools::create("/cloud/project") to create a new package. When it builds the source package, the package file will be located in the root directory of the package. However, if we use a local RStudio to create a source package, the source package will be located in the upper directory. | ||
| Line 2,068: | Line 2,327: | ||
R CMD INSTALL -l location --build pkg | R CMD INSTALL -l location --build pkg | ||
</pre> | </pre> | ||
== R folder == | |||
* https://r-pkgs.org/r.html | |||
* zzz.R. .onLoad() function. | |||
* [https://stackoverflow.com/a/20223846 R: How to run some code on load of package?] | |||
See an example from DuoClustering2018. | |||
<pre> | |||
#' @importFrom utils read.csv | |||
.onLoad <- function(libname, pkgname) { | |||
fl <- system.file("extdata", "metadata.csv", package = "DuoClustering2018") | |||
titles <- utils::read.csv(fl, stringsAsFactors = FALSE)$Title | |||
ExperimentHub::createHubAccessors(pkgname, titles) | |||
} | |||
</pre> | |||
Note that the environment of a function from the DuoClustering2018 package is not the package name. | |||
<pre> | |||
environment(clustering_summary_filteredExpr10_TrapnellTCC_v2) | |||
<environment: 0x7fe01dbe7dd0> | |||
</pre> | |||
Q: where is the definition of DuoClustering2018::clustering_summary_filteredExpr10_TrapnellTCC_v2()? A: [https://master.bioconductor.org/packages/release/bioc/vignettes/AnnotationHub/inst/doc/CreateAHubPackage.html#building-the-package createHubAccessors() - Creating A Hub Package: ExperimentHub or AnnotationHub]. | |||
== data == | == data == | ||
| Line 2,075: | Line 2,355: | ||
* If you want to store binary data and make it available to the user, put it in '''data/'''. This is the best place to put example datasets. | * If you want to store binary data and make it available to the user, put it in '''data/'''. This is the best place to put example datasets. | ||
* If you want to store parsed data, but not make it available to the user, put it in '''R/sysdata.rda'''. This is the best place to put data that your functions need. | * If you want to store parsed data, but not make it available to the user, put it in '''R/sysdata.rda'''. This is the best place to put data that your functions need. | ||
<ul> | |||
<li>If you want to store raw data, put it in '''inst/extdata'''. See [https://r-pkgs.org/data.html#data-extdata External data]. An [https://bioconductor.org/packages/release/data/experiment/vignettes/tximportData/inst/doc/tximportData.html example] from tximportData package. | |||
<pre> | |||
# grep -r extdata /home/brb/R/x86_64-pc-linux-gnu-library/4.0 | |||
logo_file <- system.file("extdata", "logo.png", package = "cowplot") | |||
orgDBLoc = system.file("extdata", "org.Hs.eg.sqlite", package="org.Hs.eg.db") | |||
# grep -r readRDS /home/brb/R/x86_64-pc-linux-gnu-library/4.0 | |||
patient.data <- readRDS("assets/coxnet.RDS") | |||
</pre> | |||
</li> | |||
</ul> | |||
[https://blog.r-hub.io/2020/05/29/distribute-data/ How to distribute data with your R package] | [https://blog.r-hub.io/2020/05/29/distribute-data/ How to distribute data with your R package] | ||
| Line 2,083: | Line 2,374: | ||
== Vignette == | == Vignette == | ||
* See examples from packages like [https://github.com/cran/magrittr magrittr] (Rmd, jpg) or [https://github.com/cran/tidyr tidyr] (Rmd, csv) or [https://github.com/cran/rvest rvest] (Rmd, png). | * [https://lifehacker.com/how-to-create-vignettes-that-really-pull-a-room-togethe-1846703615 How to Create Vignettes That Really Pull a Room Together] | ||
* See examples from packages like [https://github.com/cran/magrittr magrittr] (Rmd, jpg) or [https://github.com/cran/tidyr tidyr] (Rmd, csv) or [https://github.com/cran/rvest rvest] (Rmd, png) or [https://github.com/mikelove/DESeq2/blob/master/vignettes/DESeq2.Rmd DESeq2]. | |||
* Many vignette uses '''output: rmarkdown::html_vignette''' which will expand the width to the page. [https://github.com/mikelove/rnaseqGene/blob/master/vignettes/rnaseqGene.Rmd This one] uses '''output: BiocStyle::html_document''' which will have a [https://bioconductor.org/packages/release/workflows/vignettes/rnaseqGene/inst/doc/rnaseqGene.html narrow width]. | |||
* [http://www.markvanderloo.eu/yaRb/2019/01/11/add-a-static-pdf-vignette-to-an-r-package/ Add a static pdf vignette to an R package] | * [http://www.markvanderloo.eu/yaRb/2019/01/11/add-a-static-pdf-vignette-to-an-r-package/ Add a static pdf vignette to an R package] | ||
* [https://www.rdocumentation.org/packages/devtools/versions/2.2.0/topics/build_vignettes devtools::build_vignettes()]. The files are copied in the 'doc' directory and an vignette index is created in 'Meta/vignette.rds', as they would be in a built package. | * [https://www.rdocumentation.org/packages/devtools/versions/2.2.0/topics/build_vignettes devtools::build_vignettes()]. The files are copied in the 'doc' directory and an vignette index is created in 'Meta/vignette.rds', as they would be in a built package. | ||
| Line 2,109: | Line 2,402: | ||
[https://github.com/guangchuangyu/badger badger]: Badge for R Package | [https://github.com/guangchuangyu/badger badger]: Badge for R Package | ||
== tests folder and testthat package== | == tests folder and testthat package == | ||
* [https://www.r-bloggers.com/2025/07/the-4-layers-of-testing-every-r-package-needs/ The 4 Layers of Testing Every R Package Needs] | |||
* [http://r-pkgs.had.co.nz/tests.html testthat] package. | * [http://r-pkgs.had.co.nz/tests.html testthat] package. | ||
** [https://journal.r-project.org/archive/2011/RJ-2011-002/RJ-2011-002.pdf testthat: Get Started with Testing] 2011 | ** [https://journal.r-project.org/archive/2011/RJ-2011-002/RJ-2011-002.pdf testthat: Get Started with Testing] 2011 | ||
| Line 2,124: | Line 2,419: | ||
== .Rbuildignore == | == .Rbuildignore == | ||
[https://blog.r-hub.io/2020/05/20/rbuildignore/ Non-standard files/directories, Rbuildignore and inst] | [https://blog.r-hub.io/2020/05/20/rbuildignore/ Non-standard files/directories, Rbuildignore and inst] | ||
== URL checker == | |||
* [https://blog.r-hub.io/2020/12/01/url-checks/ A NOTE on URL checks of your R package] | |||
* [https://github.com/r-lib/urlchecker urlchecker] package | |||
== What is a library? == | == What is a library? == | ||
| Line 2,156: | Line 2,455: | ||
* [https://stackoverflow.com/questions/1454211/what-does-not-run-mean-in-r-help-pages What does "not run" mean in R help pages?] See ?example . | * [https://stackoverflow.com/questions/1454211/what-does-not-run-mean-in-r-help-pages What does "not run" mean in R help pages?] See ?example . | ||
== formatR package == | == formatR and lintr package == | ||
Use formatR package to clean up poorly formatted code | Use formatR package to clean up poorly formatted code | ||
<syntaxhighlight lang='rsplus'> | <syntaxhighlight lang='rsplus'> | ||
| Line 2,163: | Line 2,462: | ||
</syntaxhighlight> | </syntaxhighlight> | ||
Another way is to use the | Another way is to use the [https://github.com/jimhester/lintr lintr] package ([https://en.wikipedia.org/wiki/Lint_(software) lint]). | ||
<syntaxhighlight lang='rsplus'> | <syntaxhighlight lang='rsplus'> | ||
install.packages("lintr") | install.packages("lintr") | ||
| Line 2,171: | Line 2,470: | ||
== Rcpp == | == Rcpp == | ||
[https://arxiv.org/abs/1911.06416 Thirteen Simple Steps for Creating An R Package with an External C++ Library] | [https://arxiv.org/abs/1911.06416 Thirteen Simple Steps for Creating An R Package with an External C++ Library] | ||
== C library == | |||
[https://working-with-data.mazamascience.com/2021/07/16/using-r-packaging-a-c-library-in-15-minutes/ Using R — Packaging a C library in 15 minutes] | |||
== Data package == | |||
[https://rstudio4edu.github.io/rstudio4edu-book/data-pkg.html Chapter 12 Create a data package] from rstudio4edu | |||
== Minimal R package for submission == | == Minimal R package for submission == | ||
| Line 2,180: | Line 2,485: | ||
== r-hub/rhub package: the R package builder service == | == r-hub/rhub package: the R package builder service == | ||
https://github.com/r-hub/proposal | https://github.com/r-hub/proposal, [https://www.r-bloggers.com/2024/04/r-hub-v2/ R-hub v2] 2024/4. | ||
* [https://blog.r-hub.io/2019/04/08/rhub-1.1.1/ rhub 1.1.1 is on CRAN!] 2019/4/8 | * [https://blog.r-hub.io/2019/04/08/rhub-1.1.1/ rhub 1.1.1 is on CRAN!] 2019/4/8 | ||
| Line 2,297: | Line 2,602: | ||
* https://builder.r-hub.io/. See [[R#Introducing_R-hub_.28rhub_package.29.2C_the_R_package_builder_service|here]]. | * https://builder.r-hub.io/. See [[R#Introducing_R-hub_.28rhub_package.29.2C_the_R_package_builder_service|here]]. | ||
* [https://neonira.github.io/op2 The most annoying warning for CRAN submission] | * [https://neonira.github.io/op2 The most annoying warning for CRAN submission] | ||
* [https://www.mzes.uni-mannheim.de/socialsciencedatalab/article/r-package/ How to write your own R package and publish it on CRAN] '''devtools::release()''', July 2020 | |||
* [https://blog.r-hub.io/2020/12/01/url-checks/?s=09 A NOTE on URL checks of your R package] | |||
* [http://r-pkgs.had.co.nz/release.html Releasing a package] from the book "R packages" by Hadley Wickham. | |||
* [https://www.johnmackintosh.net/blog/2021-11-23-cran-success/ I got my first package onto CRAN, and YOU CAN TOO] | |||
* [https://twitter.com/PipingHotData/status/1554560631382200320?s=20&t=FSsEd2IzMdOuK1iA8iXZHg Submitting vroom to CRAN, LIVE!] | |||
=== Windows === | === Windows === | ||
[https://blog.r-hub.io/2020/04/01/win-builder/ Everything you should know about WinBuilder] | [https://blog.r-hub.io/2020/04/01/win-builder/ Everything you should know about WinBuilder] | ||
== Other tips/advice == | |||
*[https://martinctc.github.io/blog/make-package-even-more-awesome/ Top 10 tips to make your R package even more awesome] | |||
* [https://github.com/MangoTheCat/goodpractice GoodPractice] Advice on R Package Building. | |||
= C/Fortran = | = C/Fortran = | ||
== Rmath.h == | == Rmath.h == | ||
For example pnorm5() was used by [https://github.com/cran/survival/commit/3f7bbd52fc24f3fb744c5ca82946291e0735811e#diff-c5e184f4d68df5830324d0d15600667cR6 survS.h] by survival package (old version) . | For example pnorm5() was used by [https://github.com/cran/survival/commit/3f7bbd52fc24f3fb744c5ca82946291e0735811e#diff-c5e184f4d68df5830324d0d15600667cR6 survS.h] by survival package (old version) . | ||
== Makevars == | |||
* [https://cran.r-project.org/doc/manuals/R-exts.html Writing R Extensions] | |||
** The order of inclusion of makefiles for a package which does not have a src/Makefile file is ([https://cran.r-project.org/doc/manuals/R-exts.html#Package-subdirectories Section 1.1.5 Package subdirectories]) on a Unix-alike system | |||
**# src/Makevars | |||
**# R_HOME/etc/R_ARCH/Makeconf | |||
**# R_MAKEVARS_SITE, R_HOME/etc/R_ARCH/Makevars.site | |||
**# R_HOME/share/make/shlib.mk | |||
**# R_MAKEVARS_USER, ~/.R/Makevars-platform, '''~/.R/Makevars''' | |||
* [https://cran.rstudio.org/doc/manuals/R-admin.html R Installation and Administration] | |||
** [https://cran.rstudio.org/doc/manuals/R-admin.html#Customizing-package-compilation Section 6.3.3 Customizing package compilation]: This section explains that users can create a personal Makevars file to set their own compiler flags (like '''CC, CFLAGS, CXXFLAGS''', etc.). | |||
** Platform Specifics: It notes that on Unix-alikes (including macOS and Linux), the file is located at '''~/.R/Makevars'''. On Windows, it is typically found at '''Documents/.R/Makevars.win''' | |||
** Section 1.2.1 (Using Makevars): It describes how Makevars files (both inside a package's '''src/''' directory and in the user's home directory) are used to define macros for the make utility. | |||
** Variable Precedence: It explains that user-level settings in '''~/.R/Makevars''' will generally override the defaults set when R was configured, allowing you to use different compilers or include extra library paths (like the gettext path in the [[Python#Example:_benchmarKIN|benchmarKIN]] package case. | |||
== Packages includes Fortran == | == Packages includes Fortran == | ||
Some useful packages containing fortran code. | |||
* [https://github.com/cran/survC1/tree/master/src survC1] | * [https://github.com/cran/survC1/tree/master/src survC1] | ||
* [https://github.com/cran/quantreg/tree/master/src quantreg] | |||
* [https://github.com/cran/nlme/tree/master/src nlme] | * [https://github.com/cran/nlme/tree/master/src nlme] | ||
* [https://github.com/cran/glmnet/tree/master/src glmnet] | * [https://github.com/cran/glmnet/tree/master/src glmnet] | ||
* [https://github.com/cran/bsamGP/tree/master/src bsamGP] (FORTRAN 90). This is searched by using https://www.r-pkg.org/. I am not able to get what I need using the website. | * [https://github.com/cran/bsamGP/tree/master/src bsamGP] (FORTRAN 90). This is searched by using https://www.r-pkg.org/. I am not able to get what I need using the website. | ||
On mac, gfortran (6.1) can be downloaded from [https://cran.rstudio.com/bin/macosx/tools CRAN]. It will be installed onto /usr/local/gfortran. Note that the binary will not be present in PATH. So we need to run the following command to make gfortran | My experience on M1 macOS | ||
< | <ul> | ||
<li>(Outdated) On mac, gfortran (6.1) (X86 not arm) can be downloaded from [https://cran.rstudio.com/bin/macosx/tools CRAN]. It will be installed onto '''/usr/local/gfortran'''. This can be confirmed by: | |||
<pre> | |||
mkdir temp | |||
cd temp | |||
xar -xf ../gfortran-6.1.pkg | |||
lsbom Bom | |||
</pre> | |||
Note that the binary will not be present in PATH. So we need to run the following command to make gfortran availalble. | |||
<spre> | |||
sudo ln -s /usr/local/gfortran/bin/gfortran /usr/local/bin/gfortran | sudo ln -s /usr/local/gfortran/bin/gfortran /usr/local/bin/gfortran | ||
</ | </pre> | ||
<li>Tested in R 4.4.1, R is looking for '''/opt/gfortran/bin/gfortran'''. | |||
* [https://stackoverflow.com/a/77088891 Installing gfortran on MacBook with Apple M1 chip for use in R]. | |||
* See https://mac.r-project.org/tools/. Don't follow https://cran.r-project.org/bin/macosx/tools/ to download gfortran. | |||
* [https://www.r-bloggers.com/2024/01/r-universe-now-builds-macos-arm64-binaries-for-use-on-apple-silicon-aka-m1-m2-m3-systems/ R-universe now builds MacOS ARM64 binaries for use on Apple Silicon (aka M1/M2/M3) systems]. | |||
* '''Instruction''': | |||
*# Download "gfortran-12.2-universal.pkg" from https://mac.r-project.org/tools/, | |||
*# Double click the pkg file | |||
*# gfortran will be installed on "/opt/gfortran". When R installed a package containing FORTRAN code, it will automatically look for gfortran from '''/opt/gfortran/bin/gfortran'''. So it is not necessary to make gfortran available in $PATH. | |||
::[[File:GfortranMac.png|150px]] | |||
<li>Note I already had gfortran installed via homebrew for some software. But it is not used by R. | |||
<pre> | |||
$ brew uninstall gfortran | |||
Error: Refusing to uninstall /opt/homebrew/Cellar/gcc/14.1.0_1 | |||
because it is required by gstreamer, numpy, openblas and openvino, which are currently installed. | |||
You can override this and force removal with: | |||
brew uninstall --ignore-dependencies gfortran | |||
$ which -a gfortran | |||
/opt/homebrew/bin/gfortran | |||
</pre> | |||
A useful tool to find R packages containing Fortran code is [https://github.com/r-hub/pkgsearch pkgsearch] package. Note | <li>'''In order to change mess up anything''', it is better to call '''export PATH="/usr/local/bin:$PATH" ''' when we want to install R packages containing fortran code. | ||
<li>A useful tool to find R packages containing Fortran code is [https://github.com/r-hub/pkgsearch pkgsearch] package. Note | |||
* The result is not a comprehensive list of packages containing Fortran code. | * The result is not a comprehensive list of packages containing Fortran code. | ||
* It seems the result is the same as I got from https://www.r-pkg.org | * It seems the result is the same as I got from https://www.r-pkg.org | ||
| Line 2,348: | Line 2,712: | ||
20 7 lsei 1.2.0 Yong Wang 2y Solving Least Squares or... | 20 7 lsei 1.2.0 Yong Wang 2y Solving Least Squares or... | ||
</pre> | </pre> | ||
<li>Another way to find out packages containing fortran code is to run rsync to download src/contrib directory from CRAN and then use grep to find these packages. Note: the source packages takes about 8GB space (2019-10-28). | |||
Another way is to run rsync to download src/contrib directory from CRAN and then use grep to find these packages. Note: the source packages takes about 8GB space (2019-10-28). | |||
<pre> | <pre> | ||
mkdir ~/Downloads/cran | mkdir ~/Downloads/cran | ||
| Line 2,370: | Line 2,733: | ||
library(magrittr) | library(magrittr) | ||
library(stringr) | library(stringr) | ||
x <- read.table("~/Downloads/tmp", stringsAsFactors = F) | x <- read.table("~/Downloads/tmp", stringsAsFactors = F) | ||
strsplit(x$V6, "/") %>% sapply(function(x) x[1]) %>% unique() # 415 packages | strsplit(x$V6, "/") %>% sapply(function(x) x[1]) %>% unique() # 415 packages | ||
strsplit(x$V6, "/") %>% sapply(function(x) x[1]) %>% table() %>% sort() # how many Fortran files in each package | strsplit(x$V6, "/") %>% sapply(function(x) x[1]) %>% table() %>% sort() # how many Fortran files in each package | ||
str_subset(x$V6, "\\.f90$") %>% strsplit("/") %>% sapply(function(x) x[1]) %>% unique() # Fortran 90 only packages | str_subset(x$V6, "\\.f90$") %>% strsplit("/") %>% sapply(function(x) x[1]) %>% unique() # Fortran 90 only packages | ||
str_subset(x$V6, "\\.f95$") %>% strsplit("/") %>% sapply(function(x) x[1]) %>% unique() # Fortran 95 only packages | str_subset(x$V6, "\\.f95$") %>% strsplit("/") %>% sapply(function(x) x[1]) %>% unique() # Fortran 95 only packages | ||
str_subset(x$V6, "BayesFM") # f95 | str_subset(x$V6, "BayesFM") # f95 | ||
</pre> | </pre> | ||
</li> | |||
</ul> | |||
= Misc = | |||
== Datasets in R packages == | |||
https://vincentarelbundock.github.io/Rdatasets/datasets.html | |||
== Turn your analysis into a package == | |||
[https://twitter.com/neilgcurrie/status/1572254358397681665 Organise your own analysis] | |||
== Build R package faster using multicore == | |||
http://www.rexamine.com/2015/07/speeding-up-r-package-installation-process/ | |||
The idea is edit the '''/lib64/R/etc/Renviron''' file (where /lib64/R/etc/ is the result to a call to the R.home() function in R) and set: | |||
<pre> | |||
MAKE='make -j 8' # submit 8 jobs at once | |||
</pre> | |||
Then build R package as regular, for example, | |||
<pre> | |||
$ time R CMD INSTALL ~/R/stringi --preclean --configure-args='--disable-pkg-config' | |||
</pre> | |||
== suppressPackageStartupMessages() and .onAttach() == | |||
[https://github.com/cran/KernSmooth/blob/master/R/all.R KernSmooth] package example. | |||
[http://www.win-vector.com/blog/2019/08/it-is-time-for-cran-to-ban-package-ads/ It is Time for CRAN to Ban Package Ads] | |||
<syntaxhighlight lang='rsplus'> | |||
suppressPackageStartupMessages(library("dplyr")) | |||
</syntaxhighlight> | |||
== fusen package == | |||
* [https://www.r-bloggers.com/2021/08/fusen-is-now-available-on-cran/ {fusen} is now available on CRAN!] | |||
* (Videos) [https://youtu.be/febuOcS70t4?t=1742 How To Build A Package With The "Rmd First" Method], [https://youtu.be/febuOcS70t4?t=2788 fusen::inflate()] | |||
== Identifying R Functions & Packages Used in GitHub Repos == | |||
[https://www.bryanshalloway.com/2022/01/18/identifying-r-functions-packages-used-in-github-repos/ Identifying R Functions & Packages Used in GitHub Repos]: [https://brshallo.github.io/funspotr/ funspotr] package | |||
== CRANalerts == | |||
* https://cranalerts.com/ | |||
* [https://deanattali.com/blog/cranalerts/ CRANalerts: Get email alerts when a CRAN package gets updated] | |||
= | == R-universe == | ||
* [https://ropensci.org/blog/2021/06/22/setup-runiverse/ How to create your personal CRAN-like repository on R-universe] | |||
* [https://ropensci.org/blog/2023/05/31/runiverse-snapshots/ Downloading snapshots and creating stable R packages repositories using r-universe] | |||
== FDA == | |||
[https://www.r-consortium.org/blog/2021/12/08/successful-r-based-test-package-submitted-to-fda Successful R-based Test Package Submitted to FDA] | |||
== | |||
[ | |||
Latest revision as of 15:40, 18 January 2026
R package management
Challenges in Package Management
Challenges in Package Management
Packages loaded at startup
getOption("defaultPackages")
How to install a new package
- The Comprehensive Guide to Installing R Packages from CRAN, Bioconductor, GitHub and Co.
- An example from powsimR package
utils::install.packages()
- By default, install.packages() will check versions and install uninstalled packages shown in 'Depends', 'Imports' , and 'LinkingTo' (DIL) fields. See R-exts manual.
- Take advantage of Ncpus parameter in install.packages()
- By default dependencies = NA, which means c("Depends", "Imports", "LinkingTo").
- If we want to install packages listed in Suggests (or 'Enhances') field, we should specify it explicitly by using dependencies argument:
install.packages(XXXX, dependencies = c("Depends", "Imports", "Suggests", "LinkingTo")) # OR install.packages(XXXX, dependencies = TRUE)For example, if I use a plain install.packages() command to install downloader package, it only installs 'digest' and 'downloader' packages. If I useinstall.packages("downloader", dependencies=TRUE)it will also install 'testhat' package.
- Even a warning is given when some imports package is not available, it does not stop the installation (trying to install plotROC package on mac R 3.5.3). See also Warning: dependency ‘XXX’ is not available.
> install.packages("plotROC") --- Please select a CRAN mirror for use in this session --- Warning: dependency ‘gridSVG’ is not available trying URL 'https://cloud.r-project.org/bin/macosx/el-capitan/contrib/3.5/plotROC_2.2.1.tgz' Content type 'application/x-gzip' length 1288370 bytes (1.2 MB) ================================================== downloaded 1.2 MB The downloaded binary packages are in /var/folders/2q/slryb0rx4tj97t66v7l6pwvr_z6g3s/T//Rtmptrf2ZS/downloaded_packages > library(gridSVG) Error in library(gridSVG) : there is no package called ‘gridSVG’ > install.packages("gridSVG", type = "source") Warning message: package ‘gridSVG’ is not available (for R version 3.5.3) > ap <- available.packages() > "gridSVG" %in% rownames(ap) # [1] FALSE > dim(ap) # [1] 14220 17 > library(plotROC) Loading required package: ggplot2
- Look at CRAN, we see the latest version (1.7-0) of gridSVG depends on the latest version of R (3.6.0). But why even the source is removed from the old version of R?
- The install.packages function source code can be found in R -> src -> library -> utils -> R -> packages2.R file from Github repository (put 'install.packages' in the search box).
Change to use curl
Use options(download.file.method = "curl") before calling install.packages() or other install methods.
The default on Linux/MacOS is "auto" -> usually "wget" or "libcurl" if available, else "internal". When we force R to call the system's curl binary directly, bypassing wget or R's internal downloaded. Sometimes "wget" doesn't handle HTTPS well if SSL libraries aren't set up right. curl often works better with HTTPS and proxies if properly installed.
mirror
local({r <- getOption("repos")
r["CRAN"] <- "http://cran.r-project.org"
options(repos=r)
})
options("repos")
RStudio
On my mac,
- install.packages() will install the packages to User Library '/Users/USERNAME/Library/R/3.6/library'
- RStudio GUI method will install the packages to System Library '/Library/Frameworks/R.framework/Versions/3.6/Resources/library'. So if we want to remove a package, we need to specify the lib parameter in remove.packages() or clicking on the adjacent X icon to remove it.
Packages -> Install package(s) from local files...
It works on tarball file.
pacman: Install and load the package at the same time, installing temporarily
p_load() function from the pacman package.
An example of installing the purrr package.
Also used by BBC Visual and Data Journalism cookbook for R graphics
Also used by Biowulf/NIH
For bioconductor, use, for example,
pacman::p_load("BioManager")
pacman::p_load("DESeq2",try.bioconductor=TRUE,update.bioconductor=TRUE)
PS. Brute force method
mypackages<-c("ggplot2", "dplyr")
for (p in mypackages){
cond <- suppressWarnings(!require(p, character.only = TRUE))
if(cond){
try(install.packages(p), silent = TRUE)
library(p, character.only = TRUE)
}
}
remotes
devtools depends on 92 (non-base) packages and remotes depends on none.
Download and install R packages stored in 'GitHub', 'BitBucket', or plain 'subversion' or 'git' repositories. This package is a lightweight replacement of the 'install_*' functions in 'devtools'.
- https://cran.r-project.org/web/packages/remotes/index.html
- https://www.rdocumentation.org/packages/remotes/versions/2.0.2
- remotes::install("USERNAME/REPOSITORY"). Install all my packages with just a github repository and a DESCRIPTION file
To install a package from a local machines with all dependency, run remotes::install_local(path = "MyPackage.tar.gz", dependencies=TRUE) or devtools::install() though the later requires to untar the source first.
remotes::install_github() vs devtools::install_github()
See the answers. remotes is a lighter weight package for those who don't need/want all the other functionality in devtools. The remotes package says it copied most of its functionality from devtools, so the functions are likely the same.
devtools
- devtools::install()
- devtools::install(".", ...) assumes the current location contains the package content; eg Dockerfile from Bioc2020 template.
pak
- https://cran.r-project.org/web/packages/pak/index.html. 'pak' supports CRAN, 'Bioconductor' and 'GitHub' packages. An example about the censored package.
- When I use rsconnect::deployApp() to deploy a shiny app to shinyapps.io, the terminal emits the message The following package(s) were installed from sources, but may be available from the following remotes: - pak [r-lib/pak]. So it seems shinyapps.io uses the pak to install packages.
- How does this package depend on this other package?
BiocManager
https://cran.r-project.org/web/packages/BiocManager/index.html
pkgman
pkgman: A fresh approach to package installation - Gábor Csárdi
ccache: Faster R package installation
Ubuntu
- Finding Binary .deb Files for CRAN Packages
- Add an additional binary source to your source.lists file, see for example, the line in rocker. This is mentioned in Faster R package installation.
- R and CRAN Binaries for Ubuntu by Dirk Eddelbuettel
- r2u: R Binaries for Ubuntu; see The new #CRANapt repo has 19000 .deb binaries (and 200+ from BioConductor) for each with full dependencies and `apt` integration.
- rspm. {rspm}: easy access to RSPM binary packages with automatic management of system requirements
- bspm R package (cf rocker/r-rspm docker image which is not an R package). 5 lines. After that, "install.packages()" will trigger "sudo apt-get install". Note: it seems the apt-add-repository commands will break the apt update command on Debian 11. After a successful installation on Ubuntu OS, I'll see the following messages every time I started R,
Loading required package: utils Tracing function "install.packages" in package "utils"
Also for some reason, I cannot remove packages that were just installed. Not sure if the same problem happened if I use rig (based on rspm).
> install.packages("fgsea") # Works fine > remove.packages("fgsea") Removing package from ‘/home/brb/R/x86_64-pc-linux-gnu-library/4.2’ (as ‘lib’ is unspecified) Error in find.package(pkgs, lib) : there is no package called ‘fgsea’ > packageVersion("fgsea") [1] ‘1.22.0’Since the bspm::enable() function enables the integration of install_sys() into install.packages(), we have to use remove_sys(pkgs) to uninstall a package. See API.
Posit Package Manager/RStudio Package Manager/PPM, Posit Public Package Manager/P3M
- https://packagemanager.posit.co/client/#/ -> Repository:Bioconductor -> Setup.
- On Ubuntu (Linux), there is no binary format of Bioconductor packages. P3M only has binary format of CRAN packages.
- On Windows, I have to install Rtools. After Rtools has been installed, R will recognize it. Go to https://packagemanager.posit.co/client/#/. Select Bioconductor and click "SETUP". Pick Bioconductor version. Copy/paste options() statements into R or put it in the ~/.Rprofile.
- On macOS, similar to Windows.
- PPM Admin Guide.
It only supports Ubuntu (no Debian), RedHat, OpenSUSE.- In the section CRAN Binary Availability, it says Some common reasons why a package may not have a binary are: A package depends on a Bioconductor package. Package Manager does not currently support binary packages from Bioconductor, notable packages include Seurat and WGCNA.
- Posit Public Package Manager FAQ
- R Configuration Steps (Linux) -> Using Linux Binary Packages -> Configuring the R User Agent Header from Admin Guide. The step of configuring http user agent header should not be skipped.
- Posit Package Manager for Linux R Binaries. Put the following in the ~/.Rprofile file on my Debian OS. If you use the RStudio IDE, the software automatically injects this User-Agent string behind the scenes when R starts.
options(HTTPUserAgent = sprintf( "R/%s R (%s)", getRversion(), paste( getRversion(), R.version["platform"], R.version["arch"], R.version["os"] ) )) # Configure BioCManager to use Posit Package Manager: options(BioC_mirror = "https://packagemanager.posit.co/bioconductor") options(BIOCONDUCTOR_CONFIG_FILE = "https://packagemanager.posit.co/bioconductor/config.yaml") local({ # 1. Initialize with a default CRAN mirror (fallback) r <- getOption("repos") r["CRAN"] <- "https://cloud.r-project.org" # 2. Check if we are on Linux and can read os-release if (Sys.info()["sysname"] == "Linux" && file.exists("/etc/os-release")) { # Read the OS release file os_info <- readLines("/etc/os-release", warn = FALSE) # Extract the codename (e.g., "bookworm", "bullseye", "jammy") # Looks for line: VERSION_CODENAME=bookworm codename_line <- grep("^VERSION_CODENAME=", os_info, value = TRUE) if (length(codename_line) > 0) { # Remove the key and any quotes codename <- sub("^VERSION_CODENAME=\"?([^\"]+)\"?.*$", "\\1", codename_line) # 3. Construct the Posit Package Manager URL # Note: This assumes you want the 'latest' snapshot. ppm_url <- paste0("https://packagemanager.posit.co/cran/__linux__/", codename, "/latest") # Set the repo r["CRAN"] <- ppm_url } } # Apply the options options(repos = r) }) # Verify # getOption("repos") - It provides pre-compiled binary packages for Linux and Windows OS, but not macOS. When enabled, RStudio Package Manager will serve the appropriate CRAN binaries to R users instead of the source packages, saving R users significant installation time.
It does not include binary packages from bioconductor. However, Package Manager allows you to supplement local and git sources with your own precompiled binaries.- Using Bioconductor with RStudio Package Manager 7/12/2021
- See Serving Package Binaries
- It is similar to MRAN as it allows users to use particular a snapshot of CRAN as their active repositories within their R session
- programmatically retrieve frozen urls for CRAN from packagemanager.rstudio.com
- See Setup. We can change the distribution (Source, CentOS7, Rocky Linux 9, OpenSUSE, Red Hat, SLES, Ubuntu, Windows, macOS) for the Repository URL.
- See User Guide
- Once configured, users can access RStudio Package Manager through all their familiar tools including base R’s install.packages(), packrat, and devtools.
- Announcing Public Package Manager and v1.1.6
- Package Manager allows linux users to install pre-built binaries of the packages which will make install quicker. See an example here.
# Freeze to Apr 29, 2021 8:00 PM options(repos = c(REPO_NAME = "https://packagemanager.posit.co/all/2639103")) # Using Linux Binary Packages https://r-pkgs.example.com/our-cran/__linux__/xenial/latest https://packagemanager.posit.co/cran/__linux__/focal/2021-04-23
- Bring Your Own Binary Packages with RSPM
- Docker images
- r-base, rocker/r-base don't use PPM (as of R 4.3.3).
- rocker/r-ver (eg. rocker/r-ver:4.3.3) uses PPM. See the table of images on The versioned stack.
- Bioconductor (source on github) does take advantage of PPM. In Bioc 3.17/R 4.3.1, Posit's PPM hosts the binaries. As of Bioc 3.18/R 4.3.3, Bioconductor hosts the binaries.
- RStudio will be available on your web browser at http://localhost:8787. The username is "rstudio".
docker run \ -e PASSWORD=bioc \ -p 8787:8787 \ bioconductor/bioconductor_docker:devel
- R will be available on the terminal
docker run -it --rm \ --user rstudio \ bioconductor/bioconductor_docker:RELEASE_3_18 R
system.time(BiocManager::install("DESeq2", ask = FALSE)) # Replacement repositories: # CRAN: https://p3m.dev/cran/__linux__/jammy/latest # ... # trying URL 'https://bioconductor.org/packages/3.18/container-binaries/bioconductor_docker/src/contrib/bitops_1.0-7_R_x86_64-pc-linux-gnu.tar.gz' Content type 'application/gzip' length 24831 bytes (24 KB) # ... # 52 seconds
install a package as it existed on a specific date/snapshot: mran repository
- MRAN Time Machine will be retired on July 1 2023
- MRAN is getting shutdown - what else is there for reproducibility with R, or why reproducibility is on a continuum?
- MRAN snapshots, and you May 22, 2019.
Look at https://cran.microsoft.com/snapshot/, it seems the snapshot went back 2014 for R 3.1.1.
For example,
options(repos = c(CRAN = "https://mran.microsoft.com/snapshot/2022-10-01"))
install.packages("xgboost", repos="https://mran.microsoft.com/snapshot/2022-10-01/")
This trick works great when I try to install an R package (glmnet 3.0-2) on an old version of R. The current glmnet requires R 3.6.0. So even the source package is not available for older versions of R. If I install a package that does not require a very new version of R, that it works. The same problem happens on Windows OS. (today is 2020-05-05)
$ docker run --rm -it r-base:3.5.3
...
> install.packages("glmnet")
Installing package into ‘/usr/local/lib/R/site-library’
(as ‘lib’ is unspecified)
Warning message:
package ‘glmnet’ is not available (for R version 3.5.3)
> install.packages("glmnet", repos="https://mran.microsoft.com/snapshot/2019-09-20/")
...
* DONE (glmnet)
The downloaded source packages are in
‘/tmp/RtmpoWVWFQ/downloaded_packages’
> packageVersion("glmnet")
[1] ‘2.0.18’
> options()$repos
CRAN
"https://cloud.r-project.org"
> install.packages('knitr') # OK.
install a package on an old version of R
- Currently R is 4.2.2
- I have build R 4.1.3 and R 4.0.5 from source on Ubuntu 22.04. P.S. We need to include/enable "Source code" URIs Error :: You must put some 'source' URIs in your sources.list.
sudo apt update sudo apt-get build-dep r-base tar -xzvf R-4.1.3.tar.gz cd R-4.1.3 ./configure time make -j2 bin/R
- I'm trying to install DESEq2 on R 4.1.3 and R 4.0.5
- On R 4.1.3, the installation is successful.
install.packages("BiocManager") BiocManager::install("DESeq2") packageVersion("DESeq2") # 1.34.0 - On R 4.0.5, the installation of "locfit" (current version 1.5-9.7) requires R >= 4.1.0. Check on CRAN, we see the locfit version is 1.5-6 for R 4.0.5 (2021-3-31). So I try this version first.
install.packages("BiocManager") install.packages("remotes") remotes::install_version("locfit", "1.5-6") # Error: a 'NAMESPACE' file is required # Try the next version 1.5-7 remotes::install_version("locfit", "1.5-7") # Works BiocManager::install("DESeq2") # Works too packageVersion("DESeq2") # 1.30.1 - Now it is a good opportunity to try renv. So I create a new folder 'testProject' and create a new file 'test.R'. This R file contains only one line: library(DESeq2).
> setwd("testProj") > install.packages("renv") > renv::init() * Initializing project ... * Discovering package dependencies ... Done! * Copying packages into the cache ... [79/79] Done! One or more Bioconductor packages are used in your project, but the BiocManager package is not available. Consider installing BiocManager before snapshot. The following package(s) will be updated in the lockfile: ... The version of R recorded in the lockfile will be updated: - R [*] -> [4.0.5] * Lockfile written to '~/Downloads/testProject/renv.lock'. * Project '~/Downloads/testProject' loaded. [renv 0.16.0] * renv activated -- please restart the R session. > renv::install("BiocManager") # NOT SURE IF THIS IS NECESSARY Retrieving 'https://cloud.r-project.org/src/contrib/BiocManager_1.30.19.tar.gz' ... OK [file is up to date] Installing BiocManager [1.30.19] ... OK [built from source] Moving BiocManager [1.30.19] into the cache ... OK [moved to cache in 20 milliseconds] > q()Now DESeq2 is in "renv.lock" file!
Note if we accidentally start a different version of R on an renv-ed project based on an older version of R, we shall see some special message informing us The project library is out of sync with the lockfile. At this time, the "renv.lock" is not touched yet.
# Bootstrapping renv 0.16.0 -------------------------------------------------- * Downloading renv 0.16.0 ... OK (downloaded source) * Installing renv 0.16.0 ... Done! * Successfully installed and loaded renv 0.16.0. ! Using R 4.2.2 (lockfile was generated with R 4.0.5) * Project '~/Downloads/testProject' loaded. [renv 0.16.0] * The project library is out of sync with the lockfile. * Use `renv::restore()` to install packages recorded in the lockfile.
- What happened if we start with the same version of R but have a different version of renv in the global environment (vs renv.lock)?
install.packages("remotes") remotes::install_version("renv", "0.15.5") renv::init() q() # Start the same R from another directory and update renv to the latest version 0.16.0 # Now go back to the project directory and start R to see what will happen? # Nothing.... This is because the same project has the old & same version of "renv" library # in the "$PROJECT/renv" directoryIf we delete the "renv" subdirectory, ".Rprofile" file but keep "renv.lock" file, and start R (remember the 'renv' package version in the global R is different from the version in "renv.lock".
> renv::restore() This project has not yet been activated. Activating this project will ensure the project library is used during restore. Please see `?renv::activate` for more details. Would you like to activate this project before restore? [Y/n]: y Retrieving 'https://cloud.r-project.org/src/contrib/BiocManager_1.30.19.tar.gz' ... OK [file is up to date] Installing BiocManager [1.30.19] ... OK [built from source] Moving BiocManager [1.30.19] into the cache ... OK [moved to cache in 27 milliseconds] * Project '~/Downloads/testProject' loaded. [renv 0.16.0] * The project may be out of sync -- use `renv::status()` for more details. The following package(s) will be updated: # Bioconductor ======================= - AnnotationDbi [* -> 1.56.2] ... # CRAN =============================== - renv [0.16.0 -> 0.15.5] ... Do you want to proceed? [y/N]: y Retrieving 'https://cloud.r-project.org/src/contrib/Archive/renv/renv_0.15.5.tar.gz' ... OK [downloaded 961.3 Kb in 0.2 secs] Installing renv [0.15.5] ... OK [built from source] Moving renv [0.15.5] into the cache ... OK [moved to cache in 6.8 milliseconds] ... The following package(s) have been updated: renv [installed version 0.15.5 != loaded version 0.16.0] Consider restarting the R session and loading the newly-installed packages.
All the R packages are restored in the $PROJECT/renv directory.
pkgbuild: Locates compilers
https://cran.r-project.org/web/packages/pkgbuild/index.html Find Tools Needed to Build R Packages
pkgbuild::check_build_tools(debug = TRUE)
Check installed Bioconductor version
Following this post, use tools:::.BioC_version_associated_with_R_version().
Mind the '.' in front of the 'BioC'. It may be possible for some installed packages to have been sourced from a different BioC version.
tools:::.BioC_version_associated_with_R_version() # `3.6' tools:::.BioC_version_associated_with_R_version() == '3.6' # TRUE
CRAN Package Depends on Bioconductor Package
For example, if I run install.packages("NanoStringNorm") to install the package from CRAN, I may get
ERROR: dependency ‘vsn’ is not available for package ‘NanoStringNorm’
This is because the NanoStringNorm package depends on the vsn package which is on Bioconductor.
Another example is CRAN's tidyHeatmap that imports ComplexHeatmap in Bioconductor.
Another instance is CRAN's biospear (actually plsRcox) that depends on Bioc's 'survcomp' & 'mixOmics'.
One solution is to run a line setRepositories(ind=1:2). See this post or this one. Note that the default repository list can be found at (Ubuntu) /usr/lib/R/etc/repositories file.
options("repos") # display the available repositories (only CRAN)
setRepositories(ind=1:2)
options("repos") # CRAN and bioc are included
# CRAN
# "https://cloud.r-project.org"
# "https://bioconductor.org/packages/3.6/bioc"
install.packages("biospear") # it will prompt to select CRAN
install.packages("biospear", repos = "http://cran.rstudio.com") # NOT work since bioc repos is erased
This will also install the BiocInstaller package if it has not been installed before. See also Install Bioconductor Packages.
Bioconductor packages depend on CRAN
For example cowplot shows breakpointR from Bioconductor depends on it.
update.packages()
update.packages(ask="graphics") can open a graphical window to select packages.
only upgrade to binary package versions
Try setting options(pkgType = "binary") before running update.packages(). To make this always be the default behavior, you can add that line to your .Rprofile file.
How about options(install.packages.check.source = "no"), options(install.packages.compile.from.source = "never")?
Binary packages only for two versions of R
Check What Repos You are Using.
CRAN only provides binaries for one version of R prior to the current one. So when CRAN moves to post-3.6.* R most non version-stuck mirrors will not have 3.5.* binary versions of packages.
Install a tar.gz (e.g. an archived package) from a local directory
R CMD INSTALL <package-name>.tar.gz
Or in R:
# Method 1: cannot install dependencies
install.packages(<pathtopackage>, repos = NULL)
# These paths can be source directories or archives or binary package
# archive files (as created by ‘R CMD build --binary’).
# (‘http://’ and ‘file://’ URLs are also accepted and the files
# will be downloaded and installed from local copies.)
# Method 2: take care of dependencies from CRAN
devtools::install(<directory to package>, dependencies = TRUE)
# this will use 'R CMD INSTALL' to install the package.
# It will try to install dependencies of the package from CRAN,
# if they're not already installed.
The installation process can be nasty due to the dependency issue. Consider the 'biospear' package
biospear - plsRcox (archived) - plsRglm (archived) - bipartite
- lars
- pls
- kernlab
- mixOmics (CRAN->Bioconductor)
- risksetROC
- survcomp (Bioconductor)
- rms
So in order to install the 'plsRcox' package, we need to do the following steps. Note: plsRcox package is back on 6/2/2018.
# For curl
system("apt update")
system("apt install curl libcurl4-openssl-dev libssl-dev")
# For X11
system("apt install libcgal-dev libglu1-mesa-dev libglu1-mesa-dev")
system("apt install libfreetype6-dev") # https://stackoverflow.com/questions/31820865/error-in-installing-rgl-package
source("https://bioconductor.org/biocLite.R")
biocLite("survcomp") # this has to be run before the next command of installing a bunch of packages from CRAN
install.packages("https://cran.r-project.org/src/contrib/Archive/biospear/biospear_1.0.1.tar.gz",
repos = NULL, type="source")
# ERROR: dependencies ‘pkgconfig’, ‘cobs’, ‘corpcor’, ‘devtools’, ‘glmnet’, ‘grplasso’, ‘mboost’, ‘plsRcox’,
# ‘pROC’, ‘PRROC’, ‘RCurl’, ‘survAUC’ are not available for package ‘biospear’
install.packages(c("pkgconfig", "cobs", "corpcor", "devtools", "glmnet", "grplasso", "mboost",
"plsRcox", "pROC", "PRROC", "RCurl", "survAUC"))
# optional: install.packages(c("doRNG", "mvnfast"))
install.packages("https://cran.r-project.org/src/contrib/Archive/biospear/biospear_1.0.1.tar.gz",
repos = NULL, type="source")
# OR
# devtools::install_github("cran/biospear")
library(biospear) # verify
To install the (deprecated, bioc) packages 'inSilicoMerging',
biocLite(c('rjson', 'Biobase', 'RCurl'))
# destination directory is required
# download.file("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoDb_2.7.0.tar.gz",
# "~/Downloads/inSilicoDb_2.7.0.tar.gz")
# download.file("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoMerging_1.15.0.tar.gz",
# "~/Downloads/inSilicoMerging_1.15.0.tar.gz")
# ~/Downloads or $HOME/Downloads won't work in untar()
# untar("~/Downloads/inSilicoDb_2.7.0.tar.gz", exdir="/home/brb/Downloads")
# untar("~/Downloads/inSilicoMerging_1.15.0.tar.gz", exdir="/home/brb/Downloads")
# install.packages("~/Downloads/inSilicoDb", repos = NULL)
# install.packages("~/Downloads/inSilicoMerging", repos = NULL)
install.packages("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoDb_2.7.0.tar.gz",
repos = NULL, type = "source")
install.packages("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoMerging_1.15.0.tar.gz",
repos = NULL, type = "source")
R CMD INSTALL -l LIB/--library=LIB option
Install a package to a custom location
$ R CMD INSTALL -l /usr/me/localR/library myRPackage.tar.gz
Use a package installed in a custom location
R> library("myRPackage", lib.loc="/usr/me/local/R/library")
# OR include below in .bashrc file
$ export R_LIBS=/usr/me/local/R/library
R> .libPaths() # check
R> library("myRPackage")
Install a specific version of R/Bioconductor package
For packages from CRAN, use something like remotes::install_version("dplyr", "1.0.2")
For packages from Bioconductor, see the two solutions R how to install a specified version of a bioconductor package?
Install multiple/different versions of the same R package
https://stackoverflow.com/a/2989369
install.packages("~/Downloads/foo_0.1.1.tar.gz", lib = "/tmp", repos = NULL)
# a new folder "/tmp/foo" will be created
library(foo, lib.loc="/tmp") # Or use 'lib' to be consistent with install.packages()
library(foo, lib.loc="~/dev/foo/v1") ## loads v1
# and
library(foo, lib.loc="~/dev/foo/v2") ## loads v2
packageVersion("foo", lib.loc = "/tmp")
sessionInfo()
help(package = "foo", lib.loc = "/tmp")
remove.packages("foo", lib = "/tmp")
The same works for install.packages(). help(install.packages)
The install_version() from devtools and remotes will overwrite the existing installation.
Query an R package installed locally
packageDescription("MASS")
packageVersion("MASS")
Query an R package (from CRAN) basic information: available.packages()
packageStatus() # Summarize information about installed packages
available.packages() # List Available Packages at CRAN-like Repositories
# Even I use an old version of R, it still return the latest version of the packages
# The 'problem' happens on install.packages() too.
The available.packages() command is useful for understanding package dependency. Use setRepositories() or 'RGUI -> Packages -> select repositories' to select repositories and options()$repos to check or change the repositories.
The return result of available.packages() depends on the R version.
The number of packages returned from available.packages() is smaller than the number of packages obtained from rsync CRAN Mirror HOWTO/FAQ. For example, 19473 < 25000. Some archived packages are still available via rsync.
Also the packageStatus() is another useful function for query how many packages are in the repositories, how many have been installed, and individual package status (installed or not, needs to be upgraded or not).
> options()$repos
CRAN
"https://cran.rstudio.com/"
> packageStatus()
Number of installed packages:
ok upgrade unavailable
C:/Program Files/R/R-3.0.1/library 110 0 1
Number of available packages (each package counted only once):
installed not installed
http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0 76 4563
http://www.stats.ox.ac.uk/pub/RWin/bin/windows/contrib/3.0 0 5
http://www.bioconductor.org/packages/2.12/bioc/bin/windows/contrib/3.0 16 625
http://www.bioconductor.org/packages/2.12/data/annotation/bin/windows/contrib/3.0 4 686
> tmp <- available.packages()
> str(tmp)
chr [1:5975, 1:17] "A3" "ABCExtremes" "ABCp2" "ACCLMA" "ACD" "ACNE" "ADGofTest" "ADM3" "AER" ...
- attr(*, "dimnames")=List of 2
..$ : chr [1:5975] "A3" "ABCExtremes" "ABCp2" "ACCLMA" ...
..$ : chr [1:17] "Package" "Version" "Priority" "Depends" ...
> tmp[1:3,]
Package Version Priority Depends Imports LinkingTo Suggests
A3 "A3" "0.9.2" NA "xtable, pbapply" NA NA "randomForest, e1071"
ABCExtremes "ABCExtremes" "1.0" NA "SpatialExtremes, combinat" NA NA NA
ABCp2 "ABCp2" "1.1" NA "MASS" NA NA NA
Enhances License License_is_FOSS License_restricts_use OS_type Archs MD5sum NeedsCompilation File
A3 NA "GPL (>= 2)" NA NA NA NA NA NA NA
ABCExtremes NA "GPL-2" NA NA NA NA NA NA NA
ABCp2 NA "GPL-2" NA NA NA NA NA NA NA
Repository
A3 "http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0"
ABCExtremes "http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0"
ABCp2 "http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0"
And the following commands find which package depends on Rcpp and also which are from bioconductor repository.
> pkgName <- "Rcpp"
> rownames(tmp)[grep(pkgName, tmp[,"Depends"])]
> tmp[grep("Rcpp", tmp[,"Depends"]), "Depends"]
> ind <- intersect(grep(pkgName, tmp[,"Depends"]), grep("bioconductor", tmp[, "Repository"]))
> rownames(grep)[ind]
NULL
> rownames(tmp)[ind]
[1] "ddgraph" "DESeq2" "GeneNetworkBuilder" "GOSemSim" "GRENITS"
[6] "mosaics" "mzR" "pcaMethods" "Rdisop" "Risa"
[11] "rTANDEM"
CRAN vs Bioconductor packages
> R.version # 3.4.3
# CRAN
> x <- available.packages()
> dim(x)
[1] 12581 17
# Bioconductor Soft
> biocinstallRepos()
BioCsoft
"https://bioconductor.org/packages/3.6/bioc"
BioCann
"https://bioconductor.org/packages/3.6/data/annotation"
BioCexp
"https://bioconductor.org/packages/3.6/data/experiment"
CRAN
"https://cran.rstudio.com/"
> y <- available.packages(repos = biocinstallRepos()[1])
> dim(y)
[1] 1477 17
> intersect(x[, "Package"], y[, "Package"])
character(0)
# Bioconductor Annotation
> dim(available.packages(repos = biocinstallRepos()[2]))
[1] 909 17
# Bioconductor Experiment
> dim(available.packages(repos = biocinstallRepos()[3]))
[1] 326 17
# CRAN + All Bioconductor
> z <- available.packages(repos = biocinstallRepos())
> dim(z)
[1] 15292 17
Downloading Bioconductor package with an old R
When I try to download the GenomicDataCommons package using R 3.4.4 with Bioc 3.6 (the current R version is 3.5.0), it was found it can only install version 1.2.0 instead the latest version 1.4.1.
It does not work by running biocLite("BiocUpgrade") to upgrade Bioc from 3.6 to 3.7.
source("https://bioconductor.org/biocLite.R")
biocLite("BiocUpgrade")
# Error: Bioconductor version 3.6 cannot be upgraded with R version 3.4.4
See some instruction on RStudio package manager website.
Analyzing data on CRAN packages
- New undocumented function in R 3.4.0: tools::CRAN_package_db()
- http://blog.revolutionanalytics.com/2017/05/analyzing-data-on-cran-packages.html
- Six degrees of Hadley Wickham: The CRAN co-authorship network
R package location when they are installed by root
/usr/local/lib/R/site-library
Customizing your package/library location
Add a personal directory to .libPaths()
.libPaths( c( .libPaths(), "~/userLibrary") )
No need to use the assignment operator.
Install personal R packages after upgrade R, .libPaths(), Rprofile.site, R_LIBS_USER
| File | Example |
|---|---|
| Rprofile.site/.Rprofile | .libPaths(c("/usr/lib/R/site-library", "/usr/lib/R/library")) |
| Renviron.site/.Renviron | R_LIB_SITE="/usr/lib/R/site-library:/usr/lib/R/library" |
Scenario: We already have installed many R packages under R 3.1.X in the user's directory. Now we upgrade R to a new version (3.2.X). We like to have these packages available in R 3.2.X.
For Windows OS, refer to R for Windows FAQ
The follow method works on Linux and Windows.
Make sure only one instance of R is running
# Step 1. update R's built-in packages and install them on my personal directory
update.packages(ask=FALSE, checkBuilt = TRUE, repos="http://cran.rstudio.com")
# Step 2. update Bioconductor packages
.libPaths() # The first one is my personal directory
# [1] "/home/brb/R/x86_64-pc-linux-gnu-library/3.2"
# [2] "/usr/local/lib/R/site-library"
# [3] "/usr/lib/R/site-library"
# [4] "/usr/lib/R/library"
Sys.getenv("R_LIBS_USER") # may or may not equivalent to .libPaths()[1]
ul <- unlist(strsplit(Sys.getenv("R_LIBS_USER"), "/"))
src <- file.path(paste(ul[1:(length(ul)-1)], collapse="/"), "3.1")
des <- file.path(paste(ul[1:(length(ul)-1)], collapse="/"), "3.2")
pkg <- dir(src, full.names = TRUE)
if (!file.exists(des)) dir.create(des) # If 3.2 subdirectory does not exist yet!
file.copy(pkg, des, overwrite=FALSE, recursive = TRUE)
source("http://www.bioconductor.org/biocLite.R")
biocLite(ask = FALSE)
From Robert Kabacoff (R in Action)
- If you have a customized Rprofile.site file (see appendix B), save a copy outside of R.
- Launch your current version of R and issue the following statements
oldip <- installed.packages()[,1] save(oldip, file="path/installedPackages.Rdata")
where path is a directory outside of R.
- Download and install the newer version of R.
- If you saved a customized version of the Rprofile.site file in step 1, copy it into the new installation.
- Launch the new version of R, and issue the following statements
load("path/installedPackages.Rdata")
newip <- installed.packages()[,1]
for(i in setdiff(oldip, newip))
install.packages(i)
where path is the location specified in step 2.
- Delete the old installation (optional).
This approach will install only packages that are available from the CRAN. It won’t find packages obtained from other locations. In fact, the process will display a list of packages that can’t be installed For example for packages obtained from Bioconductor, use the following method to update packages
source(http://bioconductor.org/biocLite.R) biocLite(PKGNAME)
Persistent config and data for R packages with .Rprofile and .Renviron
Persistent config and data for R packages. startup, rappdirs, hoardr, keyring.
Would you like to use a personal library instead?
Some posts from internet
- Setting R_LIBS & avoiding “Would you like to use a personal library instead?”. Note: I try to create ~/.Renviron to add my personal folder in it. But update.packages() still asks me if I like to use a personal library instead (tested on Ubuntu + R 3.4).
- automatically create personal library in R. Using suppressUpdates + specify lib in biocLite() or update.packages(Sys.getenv("R_LIBS_USER"), ask = F)
# create local user library path (not present by default) dir.create(path = Sys.getenv("R_LIBS_USER"), showWarnings = FALSE, recursive = TRUE) # install to local user library path install.packages(p, lib = Sys.getenv("R_LIBS_USER"), repos = "https://cran.rstudio.com/") # Bioconductor version biocLite(p, suppressUpdates = TRUE, lib = Sys.getenv("R_LIBS_USER"))
The problem can happen if the R was installed to the C:\Program Files\R folder by users but then some main packages want to be upgraded. R will always pops a message 'Would you like to use a personal library instead?'.
To suppress the message and use the personal library always,
- Run R as administrator. If you do that, main packages can be upgraded from C:\Program Files\R\R-X.Y.Z\library folder.
- Writable R package directory cannot be found and a this. A solution here is to change the security of the R library folder so the user has a full control on the folder.
- Does R run under Windows Vista/7/8/Server 2008? There are 3 ways to get around the issue.
- I don’t have permission to write to the R-3.3.2\library directory
Actually the following hints will help us to create a convenient function UpdateMainLibrary() which will install updated main packages in the user's Documents directory without a warning dialog.
- .libPaths() only returns 1 string "C:/Program Files/R/R-x.y.z/library" on the machines that does not have this problem
- .libPaths() returns two strings "C:/Users/USERNAME/Documents/R/win-library/x.y" & "C:/Program Files/R/R-x.y.z/library" on machines with the problem.
UpdateMainLibrary <- function() {
# Update main/site packages
# The function is used to fix the problem 'Would you like to use a personal library instead?'
if (length(.libPaths()) == 1) return()
ind_mloc <- grep("Program", .libPaths()) # main library e.g. 2
ind_ploc <- grep("Documents", .libPaths()) # personal library e.g. 1
if (length(ind_mloc) > 0L && length(ind_ploc) > 0L)
# search outdated main packages
old_mloc <- ! old.packages(.libPaths()[ind_mloc])[, "Package"] %in%
installed.packages(.libPaths()[ind_ploc])[, "Package"]
oldpac <- old.packages(.libPaths()[ind_mloc])[old_mloc, "Package"]
if (length(oldpac) > 0L)
install.packages(oldpac, .libPaths()[ind_ploc])
}
On Linux,
> update.packages()
...
The downloaded source packages are in
‘/tmp/RtmpBrYccd/downloaded_packages’
Warning in install.packages(update[instlib == l, "Package"], l, contriburl = contriburl, :
'lib = "/opt/R/3.5.0/lib/R/library"' is not writable
Would you like to use a personal library instead? (yes/No/cancel) yes
...
> system("ls -lt /home/brb/R/x86_64-pc-linux-gnu-library/3.5 | head")
total 224
drwxrwxr-x 9 brb brb 4096 Oct 3 09:30 survival
drwxrwxr-x 9 brb brb 4096 Oct 3 09:29 mgcv
drwxrwxr-x 10 brb brb 4096 Oct 3 09:29 MASS
drwxrwxr-x 9 brb brb 4096 Oct 3 09:29 foreign
# So new versions of survival, mgc, MASS, foreign are installed in the personal directory
# The update.packages() will issue warnings if we try to run it again.
# It's OK to ignore these warnings.
> update.packages()
Warning: package 'foreign' in library '/opt/R/3.5.0/lib/R/library' will not be updated
Warning: package 'MASS' in library '/opt/R/3.5.0/lib/R/library' will not be updated
Warning: package 'mgcv' in library '/opt/R/3.5.0/lib/R/library' will not be updated
Warning: package 'survival' in library '/opt/R/3.5.0/lib/R/library' will not be updated
R_LIBS_USER is empty in R 3.4.1
See install.package() error, R_LIBS_USER is empty in R 3.4.1.
List vignettes from a package
vignette(package=PACKAGENAME)
List data from a package
data(package=PACKAGENAME)
sysdata.rda
List all functions of a package
Assume a package is already loaded. Then
ls("package:cowplot")
Getting a list of functions and objects in a package. This also assumes the package in loaded. In addition to functions (separated by primitive and non-primitive), it can show constants and objects.
List installed packages and versions
- http://heuristicandrew.blogspot.com/2015/06/list-of-user-installed-r-packages-and.html
- checkpoint package
ip <- as.data.frame(installed.packages()[,c(1,3:4)]) rownames(ip) <- NULL unique(ip$Priority) # [1] <NA> base recommended # Levels: base recommended ip <- ip[is.na(ip$Priority),1:2,drop=FALSE] print(ip, row.names=FALSE)
Query the names of outdated packages
psi <- packageStatus()$inst subset(psi, Status == "upgrade", drop = FALSE) # Package LibPath Version Priority Depends # RcppArmadillo RcppArmadillo C:/Users/brb/Documents/R/win-library/3.2 0.5.100.1.0 <NA> <NA> # Matrix Matrix C:/Program Files/R/R-3.2.0/library 1.2-0 recommended R (>= 2.15.2), methods # Imports LinkingTo Suggests # RcppArmadillo Rcpp (>= 0.11.0) Rcpp RUnit, Matrix, pkgKitten # Matrix graphics, grid, stats, utils, lattice <NA> expm, MASS # Enhances License License_is_FOSS License_restricts_use OS_type MD5sum # RcppArmadillo <NA> GPL (>= 2) <NA> <NA> <NA> <NA> # Matrix MatrixModels, graph, SparseM, sfsmisc GPL (>= 2) <NA> <NA> <NA> <NA> # NeedsCompilation Built Status # RcppArmadillo yes 3.2.0 upgrade # Matrix yes 3.2.0 upgrade
The above output does not show the package version from the latest packages on CRAN. So the following snippet does that.
psi <- packageStatus()$inst
pl <- unname(psi$Package[psi$Status == "upgrade"]) # List package names
ap <- as.data.frame(available.packages()[, c(1,2,3)], stringsAsFactors = FALSE)
out <- cbind(subset(psi, Status == "upgrade")[, c("Package", "Version")], ap[match(pl, ap$Package), "Version"])
colnames(out)[2:3] <- c("OldVersion", "NewVersion")
rownames(out) <- NULL
out
# Package OldVersion NewVersion
# 1 RcppArmadillo 0.5.100.1.0 0.5.200.1.0
# 2 Matrix 1.2-0 1.2-1
To consider also the packages from Bioconductor, we have the following code. Note that "3.1" means the Bioconductor version and "3.2" is the R version. See Bioconductor release versions page.
psic <- packageStatus(repos = c(contrib.url(getOption("repos")),
"http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2",
"http://www.bioconductor.org/packages/3.1/data/annotation/bin/windows/contrib/3.2"))$inst
subset(psic, Status == "upgrade", drop = FALSE)
pl <- unname(psic$Package[psic$Status == "upgrade"])
ap <- as.data.frame(available.packages(c(contrib.url(getOption("repos")),
"http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2",
"http://www.bioconductor.org/packages/3.1/data/annotation/bin/windows/contrib/3.2"))[, c(1:3)],
stringAsFactors = FALSE)
out <- cbind(subset(psic, Status == "upgrade")[, c("Package", "Version")], ap[match(pl, ap$Package), "Version"])
colnames(out)[2:3] <- c("OldVersion", "NewVersion")
rownames(out) <- NULL
out
# Package OldVersion NewVersion
# 1 limma 3.24.5 3.24.9
# 2 RcppArmadillo 0.5.100.1.0 0.5.200.1.0
# 3 Matrix 1.2-0 1.2-1
Searching for packages in CRAN
- pkgsearch package - Search R packages on CRAN
- Fishing for packages in CRAN
- CRAN now has 10,000 R packages. Here's how to find the ones you need
- Searching for R packages, packagefinder package
library(packagefinder) findPackage("survival") # 272 out of 13256 CRAN packages found in 5 seconds findPackage("follic") # No results found. # Actually 'follic' comes from randomForestSRC package # https://www.rdocumentation.org/packages/randomForestSRC/versions/2.7.0The result is shown in an html format with columns of SCORE, NAME, DESC_SHORT, DOWNL_TOTAL & GO.
METACRAN (www.r-pkg.org) - Search and browse all CRAN/R packages
- Source code on https://github.com/metacran. The 'PACKAGES' file is updated regularly to Github.
- Announcement on R/mailing list
- Author's homepage on http://gaborcsardi.org/.
cranly visualisations and summaries for R packages
Exploring R packages with cranly
Query top downloaded packages, download statistics
- Daily download statistics http://cran-logs.rstudio.com/. Note the page is split into 'package' download and 'R' download. It tracks
- Package: date, time, size, r_version, r_arch, r_os, package, version, country, ip_id.
- R: date, time, size, R version, os (win/src/osx), county, ip_id (reset daily).
- Original methods
- https://www.r-bloggers.com/finally-tracking-cran-packages-downloads/. The code still works.
- My modified code for showing the top download R packages. The original code suffers from memory issue (tested on my 64GB Linux box) when it is calling rbindlist() from data.table package with large data. Still 64GB is required since the matrix is 369M by 4 (12GB).
- Popularity bigdata / large data packages in R and ffbase useR presentation
- pkginfo: Tools for Retrieving R Package Information. It's only in github. Shiny interface.
- Top 100 downloaded packages from METACRAN
cranlogs
cranlogs package - Download Logs from the RStudio CRAN Mirror. Suitable on R console.
- 2018 through {cranlogs}
- Shiny app by Hadley (works for packages on CRAN only). It's like the Google Trend app. An example of collection: survC1, survAUC, TreatmentSelection, biospear , (glmnet)
- cranlogs 2.1.1 is on CRAN! 5/2/2019
- limit on number of packages as argument to cran_downloads
library(cranlogs)
out <- cran_top_downloads("last-week", 100) # 100 is the maximum limit
out$package
packageRank
packageRank package: Computing and Visualizing CRAN Downloads. Suitable to run on RStudio cloud. Include both CRAN and Bioconductor.
> plot(cranDownloads(packages = c("packageRank", "limma"), when = "last-month"))
> plot(cranDownloads(packages = c("shiny", "glmnet"), when = "last-month"))
> plot(cranDownloads(packages = c("shiny", "glmnet"), from = "2019", to ="2019"))
> plot(cranDownloads(packages = c("shiny", "glmnet"), from = "2019-12", to ="2019-12"))
> plot(bioconductorDownloads(packages = c("edgeR", "DESeq2", "Rsubread", "limma"), when = "last-year"))
BiocPkgTools
For Bioconductor packages, try BiocPkgTools. See the paper.
dlstats
dlstats. Monthly download stats of 'CRAN' and 'Bioconductor' packages.
Litmus: R Package Validation Reimagined
installation path not writeable from running biocLite()
When I ran biocLite() to install a new package, I got a message (the Bioc packages are installed successfully anyway)
... * DONE (curatedOvarianData) The downloaded source packages are in ‘/tmp/RtmpHxnH2K/downloaded_packages’ installation path not writeable, unable to update packages: rgl, rJava, codetools, foreign, lattice, MASS, spatial, survival
However, if I uses install.package() it can update the package
> packageVersion("survival")
[1] ‘2.42.3’
> update.packages("survival") # Not working though no error message
> packageVersion("survival")
[1] ‘2.42.3’
> install.packages("survival")
Installing package into ‘/home/brb/R/x86_64-pc-linux-gnu-library/3.4’
...
* DONE (survival)
The downloaded source packages are in
‘/tmp/RtmpHxnH2K/downloaded_packages’
> packageVersion("survival")
[1] ‘2.42.6’
> library(survival)
> sessionInfo() # show survival package 2.42-6 was attached
It makes sense to always use personal directory when we install packages. See .libPaths().
Warning: cannot remove prior installation of package
Instance 1.
# Install the latest hgu133plus2cdf package
# Remove/Uninstall hgu133plus2.db package
# Put/Install an old version of IRanges (eg version 1.18.2 while currently it is version 1.18.3)
# Test on R 3.0.1
library(hgu133plus2cdf) # hgu133pluscdf does not depend or import IRanges
source("http://bioconductor.org/biocLite.R")
biocLite("hgu133plus2.db", ask=FALSE) # hgu133plus2.db imports IRanges
# Warning:cannot remove prior installation of package 'IRanges'
# Open Windows Explorer and check IRanges folder. Only see libs subfolder.
Note:
- In the above example, all packages were installed under C:\Program Files\R\R-3.0.1\library\.
- In another instance where I cannot reproduce the problem, new R packages were installed under C:\Users\xxx\Documents\R\win-library\3.0\. The different thing is IRanges package CAN be updated but if I use packageVersion("IRanges") command in R, it still shows the old version.
- The above were tested on a desktop.
Instance 2.
# On a fresh R 3.2.0, I install Bioconductor's depPkgTools & lumi packages. Then I close R, re-open it,
# and install depPkgTools package again.
> source("http://bioconductor.org/biocLite.R")
Bioconductor version 3.1 (BiocInstaller 1.18.2), ?biocLite for help
> biocLite("pkgDepTools")
BioC_mirror: http://bioconductor.org
Using Bioconductor version 3.1 (BiocInstaller 1.18.2), R version 3.2.0.
Installing package(s) ‘pkgDepTools’
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/pkgDepTools_1.34.0.zip'
Content type 'application/zip' length 390579 bytes (381 KB)
downloaded 381 KB
package ‘pkgDepTools’ successfully unpacked and MD5 sums checked
Warning: cannot remove prior installation of package ‘pkgDepTools’
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpYd2l7i\downloaded_packages
> library(pkgDepTools)
Error in library(pkgDepTools) : there is no package called ‘pkgDepTools’
The pkgDepTools library folder appears in C:\Users\brb\Documents\R\win-library\3.2, but it is empty. The weird thing is if I try the above steps again, I cannot reproduce the problem.
Warning: dependency ‘XXX’ is not available
How should I deal with “package 'xxx' is not available (for R version x.y.z)” warning?
Error: there is no package called XXX
The error happened when I try to run library() command on a package that was just installed. R 3.6.0. 'biospear' version is 1.0.2. macOS.
> library(biospear) Loading required package: pkgconfig Error: package or namespace load failed for ‘biospear’ in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vIi): there is no package called ‘mixOmics’
Note
- The package mixOmics was removed from CRAN. It is now available on Bioconductor.
- Tested to install on a docker container: docker run --net=host -it --rm r-base
ERROR: dependency ‘car’ is not available for package ‘plsRglm’ * removing ‘/usr/local/lib/R/site-library/plsRglm’ ERROR: dependencies ‘plsRglm’, ‘mixOmics’, ‘survcomp’ are not available for package ‘plsRcox’ * removing ‘/usr/local/lib/R/site-library/plsRcox’ ERROR: dependencies ‘devtools’, ‘plsRcox’, ‘RCurl’ are not available for package ‘biospear’ * removing ‘/usr/local/lib/R/site-library/biospear’
The car package looks OK on CRAN. survcomp was moved from CRAN to Bioconductor too. - As we can see above, the official r-base image does not contain libraries enough to install RCurl/devtools packages. Consider the tidyverse image (based on RStudio image) from the rocker project.
docker pull rocker/tidyverse:3.6.0 docker run --net=host -it --rm -e PASSWORD=password -p 8787:8787 rocker/tidyverse:3.6.0 # the default username is 'rstudio' # Open a browser, log in. Run 'install.packages("RCurl")'. It works. - Testing on Mint linux also shows errors about dependencies of mixOmics and survcomp.
- The best practice to install a package that may depend on packages located/moved to Bioconductor: Run setRepositories(ind=1:2) before calling install.packages(). However, it does not remedy the situation that the 1st level imports package (eg plsRcox) was installed before but the 2nd level imports package (eg mixOmics) was not installed.
- dependsOnPkgs(): Find Reverse Dependencies. It seems it only return packages that have been installed locally. For example, tools::dependsOnPkgs("RcppEigen", "LinkingTo")
Warning: Unable to move temporary installation
The problem seems to happen only on virtual machines (Virtualbox).
- Warning: unable to move temporary installation `C:\Users\brb\Documents\R\win-library\3.0\fileed8270978f5\quadprog` to `C:\Users\brb\Documents\R\win-library\3.0\quadprog` when I try to run 'install.packages("forecast").
- Warning: unable to move temporary installation ‘C:\Users\brb\Documents\R\win-library\3.2\file5e0104b5b49\plyr’ to ‘C:\Users\brb\Documents\R\win-library\3.2\plyr’ when I try to run 'biocLite("lumi")'. The other dependency packages look fine although I am not sure if any unknown problem can happen (it does, see below).
Here is a note of my trouble shooting.
- If I try to ignore the warning and load the lumi package. I will get an error.
- If I try to run biocLite("lumi") again, it will only download & install lumi without checking missing 'plyr' package. Therefore, when I try to load the lumi package, it will give me an error again.
- Even I install the plyr package manually, library(lumi) gives another error - missing mclust package.
> biocLite("lumi")
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/BiocInstaller_1.18.2.zip'
Content type 'application/zip' length 114097 bytes (111 KB)
downloaded 111 KB
...
package ‘lumi’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages
Old packages: 'BiocParallel', 'Biostrings', 'caret', 'DESeq2', 'gdata', 'GenomicFeatures', 'gplots', 'Hmisc', 'Rcpp', 'RcppArmadillo', 'rgl',
'stringr'
Update all/some/none? [a/s/n]: a
also installing the dependencies ‘Rsamtools’, ‘GenomicAlignments’, ‘plyr’, ‘rtracklayer’, ‘gridExtra’, ‘stringi’, ‘magrittr’
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/Rsamtools_1.20.1.zip'
Content type 'application/zip' length 8138197 bytes (7.8 MB)
downloaded 7.8 MB
...
package ‘Rsamtools’ successfully unpacked and MD5 sums checked
package ‘GenomicAlignments’ successfully unpacked and MD5 sums checked
package ‘plyr’ successfully unpacked and MD5 sums checked
Warning: unable to move temporary installation ‘C:\Users\brb\Documents\R\win-library\3.2\file5e0104b5b49\plyr’
to ‘C:\Users\brb\Documents\R\win-library\3.2\plyr’
package ‘rtracklayer’ successfully unpacked and MD5 sums checked
package ‘gridExtra’ successfully unpacked and MD5 sums checked
package ‘stringi’ successfully unpacked and MD5 sums checked
package ‘magrittr’ successfully unpacked and MD5 sums checked
package ‘BiocParallel’ successfully unpacked and MD5 sums checked
package ‘Biostrings’ successfully unpacked and MD5 sums checked
Warning: cannot remove prior installation of package ‘Biostrings’
package ‘caret’ successfully unpacked and MD5 sums checked
package ‘DESeq2’ successfully unpacked and MD5 sums checked
package ‘gdata’ successfully unpacked and MD5 sums checked
package ‘GenomicFeatures’ successfully unpacked and MD5 sums checked
package ‘gplots’ successfully unpacked and MD5 sums checked
package ‘Hmisc’ successfully unpacked and MD5 sums checked
package ‘Rcpp’ successfully unpacked and MD5 sums checked
package ‘RcppArmadillo’ successfully unpacked and MD5 sums checked
package ‘rgl’ successfully unpacked and MD5 sums checked
package ‘stringr’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages
> library(lumi)
Error in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vIi) :
there is no package called ‘plyr’
Error: package or namespace load failed for ‘lumi’
> search()
[1] ".GlobalEnv" "package:BiocInstaller" "package:Biobase" "package:BiocGenerics" "package:parallel" "package:stats"
[7] "package:graphics" "package:grDevices" "package:utils" "package:datasets" "package:methods" "Autoloads"
[13] "package:base"
> biocLite("lumi")
BioC_mirror: http://bioconductor.org
Using Bioconductor version 3.1 (BiocInstaller 1.18.2), R version 3.2.0.
Installing package(s) ‘lumi’
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/lumi_2.20.1.zip'
Content type 'application/zip' length 18185326 bytes (17.3 MB)
downloaded 17.3 MB
package ‘lumi’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages
> search()
[1] ".GlobalEnv" "package:BiocInstaller" "package:Biobase" "package:BiocGenerics" "package:parallel" "package:stats"
[7] "package:graphics" "package:grDevices" "package:utils" "package:datasets" "package:methods" "Autoloads"
[13] "package:base"
> library(lumi)
Error in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vIi) :
there is no package called ‘plyr’
Error: package or namespace load failed for ‘lumi’
> biocLite("plyr")
BioC_mirror: http://bioconductor.org
Using Bioconductor version 3.1 (BiocInstaller 1.18.2), R version 3.2.0.
Installing package(s) ‘plyr’
trying URL 'http://cran.rstudio.com/bin/windows/contrib/3.2/plyr_1.8.2.zip'
Content type 'application/zip' length 1128621 bytes (1.1 MB)
downloaded 1.1 MB
package ‘plyr’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages
> library(lumi)
Error in loadNamespace(j <- i1L, c(lib.loc, .libPaths()), versionCheck = vIj) :
there is no package called ‘mclust’
Error: package or namespace load failed for ‘lumi’
> ?biocLite
Warning messages:
1: In read.dcf(file.path(p, "DESCRIPTION"), c("Package", "Version")) :
cannot open compressed file 'C:/Users/brb/Documents/R/win-library/3.2/Biostrings/DESCRIPTION', probable reason 'No such file or directory'
2: In find.package(if (is.null(package)) loadedNamespaces() else package, :
there is no package called ‘Biostrings’
> library(lumi)
Error in loadNamespace(j <- i1L, c(lib.loc, .libPaths()), versionCheck = vIj) :
there is no package called ‘mclust’
In addition: Warning messages:
1: In read.dcf(file.path(p, "DESCRIPTION"), c("Package", "Version")) :
cannot open compressed file 'C:/Users/brb/Documents/R/win-library/3.2/Biostrings/DESCRIPTION', probable reason 'No such file or directory'
2: In find.package(if (is.null(package)) loadedNamespaces() else package, :
there is no package called ‘Biostrings’
Error: package or namespace load failed for ‘lumi’
Other people also have the similar problem. The possible cause is the virus scanner locks the file and R cannot move them.
Some possible solutions:
- Delete ALL folders under R/library (e.g. C:/Progra~1/R/R-3.2.0/library) folder and install the main package again using install.packages() or biocLite().
- For specific package like 'lumi' from Bioconductor, we can find out all dependency packages and then install them one by one.
- Find out and install the top level package which misses dependency packages.
- This is based on the fact that install.packages() or biocLite() sometimes just checks & installs the 'Depends' and 'Imports' packages and won't install all packages recursively
- we can do a small experiment by removing a package which is not directly dependent/imported by another package; e.g. 'iterators' is not dependent/imported by 'glment' directly but indirectly. So if we run remove.packages("iterators"); install.packages("glmnet"), then the 'iterator' package is still missing.
- A real example is if the missing packages are 'Biostrings', 'limma', 'mclust' (these are packages that 'minfi' directly depends/imports although they should be installed when I run biocLite("lumi") command), then I should just run the command remove.packages("minfi"); biocLite("minfi"). If we just run biocLite("lumi") or biocLite("methylumi"), the missing packages won't be installed.
Error in download.file(url, destfile, method, mode = "wb", ...)
HTTP status was '404 Not Found'
Tested on an existing R-3.2.0 session. Note that VariantAnnotation 1.14.4 was just uploaded to Bioc.
> biocLite("COSMIC.67")
BioC_mirror: http://bioconductor.org
Using Bioconductor version 3.1 (BiocInstaller 1.18.3), R version 3.2.0.
Installing package(s) ‘COSMIC.67’
also installing the dependency ‘VariantAnnotation’
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/VariantAnnotation_1.14.3.zip'
Error in download.file(url, destfile, method, mode = "wb", ...) :
cannot open URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/VariantAnnotation_1.14.3.zip'
In addition: Warning message:
In download.file(url, destfile, method, mode = "wb", ...) :
cannot open: HTTP status was '404 Not Found'
Warning in download.packages(pkgs, destdir = tmpd, available = available, :
download of package ‘VariantAnnotation’ failed
installing the source package ‘COSMIC.67’
trying URL 'http://bioconductor.org/packages/3.1/data/experiment/src/contrib/COSMIC.67_1.4.0.tar.gz'
Content type 'application/x-gzip' length 40999037 bytes (39.1 MB)
However, when I tested on a new R-3.2.0 (just installed in VM), the COSMIC package installation is successful. That VariantAnnotation version 1.14.4 was installed (this version was just updated today from Bioconductor).
The cause of the error is the available.package() function will read the rds file first from cache in a tempdir (C:\Users\XXXX\AppData\Local\Temp\RtmpYYYYYY). See lines 51-55 of <packages.R>.
dest <- file.path(tempdir(),
paste0("repos_", URLencode(repos, TRUE), ".rds"))
if(file.exists(dest)) {
res0 <- readRDS(dest)
} else {
...
}
Since my R was opened 1 week ago, the rds file it reads today contains old information. Note that Bioconductor does not hold the source code or binary code for the old version of packages. This explains why biocLite() function broke. When I restart R, the original problem is gone.
If we look at the source code of available.packages(), we will see we could use cacheOK option in download.file() function.
download.file(url, destfile, method, cacheOK = FALSE, quiet = TRUE, mode ="wb")
Another case: Error in download.file(url, destfile, method, mode = "wb", ...)
> install.packages("quantreg")
There is a binary version available but the source version is later:
binary source needs_compilation
quantreg 5.33 5.34 TRUE
Do you want to install from sources the package which needs compilation?
y/n: n
trying URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.4/quantreg_5.33.tgz'
Warning in install.packages :
cannot open URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.4/quantreg_5.33.tgz': HTTP status was '404 Not Found'
Error in download.file(url, destfile, method, mode = "wb", ...) :
cannot open URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.4/quantreg_5.33.tgz'
Warning in install.packages :
download of package ‘quantreg’ failed
It seems the binary package cannot be found on the mirror. So the solution here is to download the package from the R main server. Note that after I have successfully installed the binary package from the main R server, I remove the package in R and try to install the binary package from rstudio.com server agin and it works this time.
> install.packages("quantreg", repos = "https://cran.r-project.org")
trying URL 'https://cran.r-project.org/bin/macosx/el-capitan/contrib/3.4/quantreg_5.34.tgz'
Content type 'application/x-gzip' length 1863561 bytes (1.8 MB)
==================================================
downloaded 1.8 MB
Another case: Error in download.file() on Windows 7
For some reason, IE 8 cannot interpret https://ftp.ncbi.nlm.nih.gov though it understands ftp://ftp.ncbi.nlm.nih.gov.
This is tested using R 3.4.3.
> download.file("https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", "test.soft.gz")
trying URL 'https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz'
Error in download.file("https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", :
cannot open URL 'https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz'
In addition: Warning message:
In download.file("https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", :
InternetOpenUrl failed: 'An error occurred in the secure channel support'
> download.file("ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", "test.soft.gz")
trying URL 'ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz'
downloaded 9.1 MB
ERROR: failed to lock directory
Follow the suggestion to remove the LOCK file. See the post.
It could happened in calling install.packages(), biocLite() or devtools::install_github(), and so on.
Error in unloadNamespace(package)
> d3heatmap(mtcars, scale = "column", colors = "Blues")
Error: 'col_numeric' is not an exported object from 'namespace:scales'
> packageVersion("scales")
[1] ‘0.2.5’
> library(scales)
Error in unloadNamespace(package) :
namespace ‘scales’ is imported by ‘ggplot2’ so cannot be unloaded
In addition: Warning message:
package ‘scales’ was built under R version 3.2.1
Error in library(scales) :
Package ‘scales’ version 0.2.4 cannot be unloaded
> search()
[1] ".GlobalEnv" "package:d3heatmap" "package:ggplot2"
[4] "package:microbenchmark" "package:COSMIC.67" "package:BiocInstaller"
[7] "package:stats" "package:graphics" "package:grDevices"
[10] "package:utils" "package:datasets" "package:methods"
[13] "Autoloads" "package:base"
If I open a new R session, the above error will not happen!
The problem occurred because the 'scales' package version required by the d3heatmap package/function is old. See this post. And when I upgraded the 'scales' package, it was locked by the package was imported by the ggplot2 package.
Unload a package
Add unload = TRUE option to unload the namespace. See detach().
require(splines) detach(package:splines, unload=TRUE)
crantastic
https://crantastic.org/. A community site for R packages where you can search for, review and tag CRAN packages.
https://github.com/hadley/crantastic
New R packages as reported by CRANberries
http://blog.revolutionanalytics.com/2015/07/mranspackages-spotlight.html
#----------------------------
# SCRAPE CRANBERRIES FILES TO COUNT NEW PACKAGES AND PLOT
#
library(ggplot2)
# Build a vextor of the directories of interest
year <- c("2013","2014","2015")
month <- c("01","02","03","04","05","06","07","08","09","10","11","12")
span <-c(rep(month,2),month[1:7])
dir <- "http://dirk.eddelbuettel.com/cranberries"
url2013 <- file.path(dir,"2013",month)
url2014 <- file.path(dir,"2014",month)
url2015 <- file.path(dir,"2015",month[1:7])
url <- c(url2013,url2014,url2015)
# Read each directory and count the new packages
new_p <- vector()
for(i in url){
raw.data <- readLines(i)
new_p[i] <- length(grep("New package",raw.data,value=TRUE))
}
# Plot
time <- seq(as.Date("2013-01-01"), as.Date("2015-07-01"), by="months")
new_pkgs <- data.frame(time,new_p)
ggplot(new_pkgs, aes(time,y=new_p)) +
geom_line() + xlab("") + ylab("Number of new packages") +
geom_smooth(method='lm') + ggtitle("New R packages as reported by CRANberries")
R packages being removed
- https://dirk.eddelbuettel.com/cranberries/cran/removed/
- https://web.archive.org/ to find out the package website before it's removed
Top new packages in 2015
- 2015 R packages roundup by CHRISTOPH SAFFERLING
- R trends in 2015 by MAX GORDON
keep.source.pkgs option
State of R packages in your library. The original code formatting and commenting being removed by default, unless one changes some options for installing packages.
- options(keep.source.pkgs = TRUE)
- install.packages("rhub", INSTALL_opts = "--with-keep.source", type = "source")
- R CMD install --with-keep.source
Package installation speed for packages installed with ‘keep.source’ has been improved. 2021-12-2.
Speeding up package installation
- http://blog.jumpingrivers.com/posts/2017/speed_package_installation/
- (Much) Faster Package (Re-)Installation via Caching
- (Much) Faster Package (Re-)Installation via Caching, part 2
An efficient way to install and load R packages
An efficient way to install and load R packages
# Package names
packages <- c("ggplot2", "readxl", "dplyr", "tidyr", "ggfortify", "DT", "reshape2", "knitr", "lubridate", "pwr", "psy", "car", "doBy", "imputeMissings", "RcmdrMisc", "questionr", "vcd", "multcomp", "KappaGUI", "rcompanion", "FactoMineR", "factoextra", "corrplot", "ltm", "goeveg", "corrplot", "FSA", "MASS", "scales", "nlme", "psych", "ordinal", "lmtest", "ggpubr", "dslabs", "stringr", "assist", "ggstatsplot", "forcats", "styler", "remedy", "snakecaser", "addinslist", "esquisse", "here", "summarytools", "magrittr", "tidyverse", "funModeling", "pander", "cluster", "abind")
# Install packages not yet installed
installed_packages <- packages %in% rownames(installed.packages())
if (any(installed_packages == FALSE)) {
install.packages(packages[!installed_packages])
}
# Packages loading
invisible(lapply(packages, library, character.only = TRUE))
Alternatively use the pacman package.
library( , exclude, include.only)
See ?library
library(tidyverse) library(MASS, exclude='select') library(thepackage, include.only="thefunction")
package ‘XXX’ was installed by an R version with different internals
it needs to be reinstalled for use with this R version. The problem seems to be specific to R 3.5.1 in Ubuntu 16.04. I got this message when I try to install the keras and tidyverse packages. The 'XXX' package includes nlme for installing "tidyverse" and Matrix for installing "reticulate". I have already logged in as root. I need to manually install these packages again though it seems I did not see a version change for these packages.
Today it also happened when I tried to install "pec" which broke when it was installing "Hmisc". The error message is "Error : package ‘rpart’ was installed by an R version with different internals; it needs to be reinstalled for use with this R version". I am using R 3.5.2. rpart version is ‘4.1.13’. The solution is I install rpart again (under my account is enough) though rpart does not have a new version. Then I can install "Hmisc".
packrat and renv
See Reproducible → packrat/renv
R package dependencies
Depends, Imports, Suggests, Enhances, LinkingTo
See Writing R Extensions and install.packages().
- Depends: list of package names which this package depends on. Those packages will be attached (so it is better to use Imports instead of Depends as much as you can) before the current package when library or require is called. The ‘Depends’ field can also specify a dependence on a certain version of R.
- Imports: lists packages whose namespaces are imported from (as specified in the NAMESPACE file) but which do not need to be attached.
- Suggests: lists packages that are not necessarily needed. This includes packages used only in examples, tests or vignettes, and packages loaded in the body of functions.
- Enhances: lists packages “enhanced” by the package at hand, e.g., by providing methods for classes from these packages, or ways to handle objects from these packages.
- LinkingTo: A package that wishes to make use of header files in other packages needs to declare them as a comma-separated list in the field ‘LinkingTo’ in the DESCRIPTION file.
- An example is SingleR that links to the beachmat package in its cpp source code.
- available.packages(); see packageStatus().
- download.packages()
- packageStatus(), update(), upgrade(). packageStatus() will return a list with two components:
- inst - a data frame with columns as the matrix returned by installed.packages plus "Status", a factor with levels c("ok", "upgrade"). Note: the manual does not mention "unavailable" case (but I do get it) in R 3.2.0?
- avail - a data frame with columns as the matrix returned by available.packages plus "Status", a factor with levels c("installed", "not installed", "unavailable"). Note: I don't get the "unavailable" case in R 3.2.0?
> x <- packageStatus() > names(x) [1] "inst" "avail" > dim(x'inst') [1] 225 17 > x'inst'[1:3, ] Package LibPath Version Priority Depends Imports acepack acepack C:/Program Files/R/R-3.1.2/library 1.3-3.3 <NA> <NA> <NA> adabag adabag C:/Program Files/R/R-3.1.2/library 4.0 <NA> rpart, mlbench, caret <NA> affxparser affxparser C:/Program Files/R/R-3.1.2/library 1.38.0 <NA> R (>= 2.6.0) <NA> LinkingTo Suggests Enhances acepack <NA> <NA> <NA> adabag <NA> <NA> <NA> affxparser <NA> R.oo (>= 1.18.0), R.utils (>= 1.32.4),\nAffymetrixDataTestFiles <NA> License License_is_FOSS License_restricts_use OS_type MD5sum NeedsCompilation Built acepack MIT + file LICENSE <NA> <NA> <NA> <NA> yes 3.1.2 adabag GPL (>= 2) <NA> <NA> <NA> <NA> no 3.1.2 affxparser LGPL (>= 2) <NA> <NA> <NA> <NA> <NA> 3.1.1 Status acepack ok adabag ok affxparser unavailable > dim(x'avail') [1] 6538 18 > x'avail'[1:3, ] Package Version Priority Depends Imports LinkingTo A3 A3 0.9.2 <NA> R (>= 2.15.0), xtable, pbapply <NA> <NA> ABCExtremes ABCExtremes 1.0 <NA> SpatialExtremes, combinat <NA> <NA> ABCanalysis ABCanalysis 1.0.1 <NA> R (>= 2.10) Hmisc, plotrix <NA> Suggests Enhances License License_is_FOSS License_restricts_use OS_type Archs A3 randomForest, e1071 <NA> GPL (>= 2) <NA> <NA> <NA> <NA> ABCExtremes <NA> <NA> GPL-2 <NA> <NA> <NA> <NA> ABCanalysis <NA> <NA> GPL-3 <NA> <NA> <NA> <NA> MD5sum NeedsCompilation File Repository Status A3 <NA> <NA> <NA> http://cran.rstudio.com/bin/windows/contrib/3.1 not installed ABCExtremes <NA> <NA> <NA> http://cran.rstudio.com/bin/windows/contrib/3.1 not installed ABCanalysis <NA> <NA> <NA> http://cran.rstudio.com/bin/windows/contrib/3.1 not installed
- packageVersion(), packageDescription()
- install.packages(), remove.packages().
- installed.packages(); see packageStatus().
- update.packages(), old.packages(), new.packages()
- setRepositories()
- contrib.url()
- chooseCRANmirror(), chooseBioCmirror()
- suppressForeignCheck()
download.packages() for source package
Consider the esimate package hosted on r-forge.
download.packages("estimate", destdir = "~/Downloads",
repos = "https://R-Forge.R-project.org")
tools package
- https://www.rdocumentation.org/packages/tools/versions/3.6.1
- CRAN_package_db() from ?CRANtools. Especially, it gives the Description and Maintainer information not provided by utils::available.packages()
- dependsOnPkgs()
db <- tools::CRAN_package_db() nRcpp <- length(tools::dependsOnPkgs("Rcpp", recursive=FALSE, installed=db) ) nCompiled <- table(db[, "NeedsCompilation"])[["yes"]] propRcpp <- nRcpp / nCompiled * 100
- package.dependencies(), pkgDepends(), etc are deprecated now, mostly in favor of package_dependencies() which is both more flexible and efficient. See R 3.3.0 News. For example, tools::package_dependencies(c("remotes", "devtools"), recursive=TRUE) shows remotes has only a few dependencies while devtools has a lot.
crandep package
https://cran.r-project.org/web/packages/crandep/index.html. Useful to find reverse dependencies. ?get_dep. Consider the abc package:
get_dep("abc", "depends") # abc depends on these packages
# note my computer does not have 'abc' installed
# from to type reverse
# 1 abc abc.data depends FALSE
# 2 abc nnet depends FALSE
# 3 abc quantreg depends FALSE
# 4 abc MASS depends FALSE
# 5 abc locfit depends FALSE
get_dep("abc", "reverse_depends")
# from to type reverse
# 1 abc abctools depends TRUE
# 2 abc EasyABC depends TRUE
x <- get_dep("RcppEigen", c("reverse linking to"))
dim(x)
# [1] 331 4
head(x, 3)
# from to type reverse
# 1 RcppEigen abess linking to TRUE
# 2 RcppEigen acrt linking to TRUE
# 3 RcppEigen ADMMnet linking to TRUE
How does this package depend on this other package
How does this package depend on this other package?, pak::pkg_deps_explain()
pkgndep
- CRAN & the paper pkgndep: a tool for analyzing dependency heaviness of R packages
- Package dependencies in your session. DESeq2 was used.
remotes
remotes::local_package_deps(dependencies=TRUE) will find and return all dependent packages based on the "DESCRIPTION" file. See an example here.
Bioconductor's pkgDepTools package
The is an example of querying the dependencies of the notorious 'lumi' package which often broke the installation script. I am using R 3.2.0 and Bioconductor 3.1.
The getInstallOrder function is useful to get a list of all (recursive) dependency packages.
source("http://bioconductor.org/biocLite.R")
if (!require(pkgDepTools)) {
biocLite("pkgDepTools", ask = FALSE)
library(pkgDepTools)
}
MkPlot <- FALSE
library(BiocInstaller)
biocUrl <- biocinstallRepos()["BioCsoft"]
biocDeps <- makeDepGraph(biocUrl, type="source", dosize=FALSE) # pkgDepTools defines its makeDepGraph()
PKG <- "lumi"
if (MkPlot) {
if (!require(Biobase)) {
biocLite("Biobase", ask = FALSE)
library(Biobase)
}
if (!require(Rgraphviz)) {
biocLite("Rgraphviz", ask = FALSE)
library(Rgraphviz)
}
categoryNodes <- c(PKG, names(acc(biocDeps, PKG)1))
categoryGraph <- subGraph(categoryNodes, biocDeps)
nn <- makeNodeAttrs(categoryGraph, shape="ellipse")
plot(categoryGraph, nodeAttrs=nn) # Complete but plot is too complicated & font is too small.
}
system.time(allDeps <- makeDepGraph(biocinstallRepos(), type="source",
keep.builtin=TRUE, dosize=FALSE)) # takes a little while
# user system elapsed
# 175.737 10.994 186.875
# Warning messages:
# 1: In .local(from, to, graph) : edges replaced: ‘SNPRelate|gdsfmt’
# 2: In .local(from, to, graph) :
# edges replaced: ‘RCurl|methods’, ‘NA|bitops’
# When needed.only=TRUE, only those dependencies not currently installed are included in the list.
x1 <- sort(getInstallOrder(PKG, allDeps, needed.only=TRUE)$packages); x1
[1] "affy" "affyio"
[3] "annotate" "AnnotationDbi"
[5] "base64" "beanplot"
[7] "Biobase" "BiocParallel"
[9] "biomaRt" "Biostrings"
[11] "bitops" "bumphunter"
[13] "colorspace" "DBI"
[15] "dichromat" "digest"
[17] "doRNG" "FDb.InfiniumMethylation.hg19"
[19] "foreach" "futile.logger"
[21] "futile.options" "genefilter"
[23] "GenomeInfoDb" "GenomicAlignments"
[25] "GenomicFeatures" "GenomicRanges"
[27] "GEOquery" "ggplot2"
[29] "gtable" "illuminaio"
[31] "IRanges" "iterators"
[33] "labeling" "lambda.r"
[35] "limma" "locfit"
[37] "lumi" "magrittr"
[39] "matrixStats" "mclust"
[41] "methylumi" "minfi"
[43] "multtest" "munsell"
[45] "nleqslv" "nor1mix"
[47] "org.Hs.eg.db" "pkgmaker"
[49] "plyr" "preprocessCore"
[51] "proto" "quadprog"
[53] "RColorBrewer" "Rcpp"
[55] "RCurl" "registry"
[57] "reshape" "reshape2"
[59] "rngtools" "Rsamtools"
[61] "RSQLite" "rtracklayer"
[63] "S4Vectors" "scales"
[65] "siggenes" "snow"
[67] "stringi" "stringr"
[69] "TxDb.Hsapiens.UCSC.hg19.knownGene" "XML"
[71] "xtable" "XVector"
[73] "zlibbioc"
# When needed.only=FALSE the complete list of dependencies is given regardless of the set of currently installed packages.
x2 <- sort(getInstallOrder(PKG, allDeps, needed.only=FALSE)$packages); x2
[1] "affy" "affyio" "annotate"
[4] "AnnotationDbi" "base64" "beanplot"
[7] "Biobase" "BiocGenerics" "BiocInstaller"
[10] "BiocParallel" "biomaRt" "Biostrings"
[13] "bitops" "bumphunter" "codetools"
[16] "colorspace" "DBI" "dichromat"
[19] "digest" "doRNG" "FDb.InfiniumMethylation.hg19"
[22] "foreach" "futile.logger" "futile.options"
[25] "genefilter" "GenomeInfoDb" "GenomicAlignments"
[28] "GenomicFeatures" "GenomicRanges" "GEOquery"
[31] "ggplot2" "graphics" "grDevices"
[34] "grid" "gtable" "illuminaio"
[37] "IRanges" "iterators" "KernSmooth"
[40] "labeling" "lambda.r" "lattice"
[43] "limma" "locfit" "lumi"
[46] "magrittr" "MASS" "Matrix"
[49] "matrixStats" "mclust" "methods"
[52] "methylumi" "mgcv" "minfi"
[55] "multtest" "munsell" "nleqslv"
[58] "nlme" "nor1mix" "org.Hs.eg.db"
[61] "parallel" "pkgmaker" "plyr"
[64] "preprocessCore" "proto" "quadprog"
[67] "RColorBrewer" "Rcpp" "RCurl"
[70] "registry" "reshape" "reshape2"
[73] "rngtools" "Rsamtools" "RSQLite"
[76] "rtracklayer" "S4Vectors" "scales"
[79] "siggenes" "snow" "splines"
[82] "stats" "stats4" "stringi"
[85] "stringr" "survival" "tools"
[88] "TxDb.Hsapiens.UCSC.hg19.knownGene" "utils" "XML"
[91] "xtable" "XVector" "zlibbioc"
> sort(setdiff(x2, x1)) # Not all R's base packages are included; e.g. 'base', 'boot', ...
[1] "BiocGenerics" "BiocInstaller" "codetools" "graphics" "grDevices"
[6] "grid" "KernSmooth" "lattice" "MASS" "Matrix"
[11] "methods" "mgcv" "nlme" "parallel" "splines"
[16] "stats" "stats4" "survival" "tools" "utils"
Bioconductor BiocPkgTools
Collection of simple tools for learning about Bioc Packages. Functionality includes access to :
- Download statistics
- General package listing
- Build reports
- Package dependency graphs
- Vignettes
Overview of BiocPkgTools & Dependency graphs
BiocPkgTools: Toolkit for Mining the Bioconductor Package Ecosystem in biorxiv.org.
miniCRAN package
miniCRAN package can be used to identify package dependencies or create a local CRAN repository. It can be used on repositories other than CRAN, such as Bioconductor.
- http://blog.revolutionanalytics.com/2014/07/dependencies-of-popular-r-packages.html
- http://www.r-bloggers.com/introducing-minicran-an-r-package-to-create-a-private-cran-repository/
- http://www.magesblog.com/2014/09/managing-r-package-dependencies.html
- Using miniCRAN in Azure ML
- developing internal CRAN Repositories
Before we go into R, we need to install some packages from Ubuntu terminal. See here.
# Consider glmnet package (today is 4/29/2015)
# Version: 2.0-2
# Depends: Matrix (≥ 1.0-6), utils, foreach
# Suggests: survival, knitr, lars
if (!require("miniCRAN")) {
install.packages("miniCRAN", dependencies = TRUE, repos="http://cran.rstudio.com") # include 'igraph' in Suggests.
library(miniCRAN)
}
if (!"igraph" %in% installed.packages()[,1]) install.packages("igraph")
tags <- "glmnet"
pkgDep(tags, suggests=TRUE, enhances=TRUE) # same as pkgDep(tags)
# [1] "glmnet" "Matrix" "foreach" "codetools" "iterators" "lattice" "evaluate" "digest"
# [9] "formatR" "highr" "markdown" "stringr" "yaml" "mime" "survival" "knitr"
# [17] "lars"
dg <- makeDepGraph(tags, suggests=TRUE, enhances=TRUE) # miniCRAN defines its makeDepGraph()
plot(dg, legendPosition = c(-1, 1), vertex.size=20)
We can also display the dependence for a package from the Bioconductor repository.
tags <- "DESeq2"
# Depends S4Vectors, IRanges, GenomicRanges, Rcpp (>= 0.10.1), RcppArmadillo (>= 0.3.4.4)
# Imports BiocGenerics(>= 0.7.5), Biobase, BiocParallel, genefilter, methods, locfit, geneplotter, ggplot2, Hmisc
# Suggests RUnit, gplots, knitr, RColorBrewer, BiocStyle, airway,\npasilla (>= 0.2.10), DESeq, vsn
# LinkingTo Rcpp, RcppArmadillo
index <- function(url, type="source", filters=NULL, head=5, cols=c("Package", "Version")){
contribUrl <- contrib.url(url, type=type)
available.packages(contribUrl, type=type, filters=filters)
}
bioc <- local({
env <- new.env()
on.exit(rm(env))
evalq(source("http://bioconductor.org/biocLite.R", local=TRUE), env)
biocinstallRepos() # return URLs
})
bioc
# BioCsoft
# "http://bioconductor.org/packages/3.0/bioc"
# BioCann
# "http://bioconductor.org/packages/3.0/data/annotation"
# BioCexp
# "http://bioconductor.org/packages/3.0/data/experiment"
# BioCextra
# "http://bioconductor.org/packages/3.0/extra"
# CRAN
# "http://cran.fhcrc.org"
# CRANextra
# "http://www.stats.ox.ac.uk/pub/RWin"
str(index(bioc["BioCsoft"])) # similar to cranJuly2014 object
system.time(dg <- makeDepGraph(tags, suggests=TRUE, enhances=TRUE, availPkgs = index(bioc["BioCsoft"]))) # Very quick!
plot(dg, legendPosition = c(-1, 1), vertex.size=20)
The dependencies of GenomicFeature and GenomicAlignments are more complicated. So we turn the 'suggests' option to FALSE.
tags <- "GenomicAlignments" dg <- makeDepGraph(tags, suggests=FALSE, enhances=FALSE, availPkgs = index(bioc["BioCsoft"])) plot(dg, legendPosition = c(-1, 1), vertex.size=20)
Github repository
cranlike
https://github.com/r-hub/cranlike
cranlike keeps the package data in a SQLite database, in addition to the PACKAGES* files. This database is the canonical source of the package data. It can be updated quickly, to add and remove packages. The PACKAGES* files are generated from the database.
MRAN (CRAN only) & checkpoint package
- http://blog.revolutionanalytics.com/2014/10/explore-r-package-connections-at-mran.html
- Reproducible Work in R
- MRAN is Dead, long live GRAN 4/28/2023
According to the snapsot list here, the oldest version is 2014-08-18 which corresponds to R 3.1.0.
checkpoint package
library(checkpoint)
checkpoint("2015-03-31")
Note the Bioconductor packages have no similar solution.
groundhog package
- groundhog: Version-Control for CRAN, GitHub, and GitLab Packages (Bioconductor?)
- It seems groundhog is like checkpoint but groundhog does not depend on MRAN and it will figure out the package dependencies by itself.
- MRAN is getting shutdown - what else is there for reproducibility with R, or why reproducibility is on a continuum? The author provides an example where we can integrate "groundhog" in the Dockerfile for reproducibility. Pay attention to the sentence (appear 2 times) “why use Docker at all? Since it’s easy to install older versions of R on Windows and macOS, wouldn’t an renv.lock file suffice? Or even just {groundhog} which is arguably even easier to use?”
- MRAN is getting shutdown #593 related to Rocker project. Rocker now uses RSPM (PPM now) from RStudio.
- groundhog.library() differs from the library() function: 1) it installed and loaded packages in one step, 2) it allowed to install multiple packages.
Note I use the docker's R since that's the easiest way to use an old version of R (e.g. for some old R script) in Linux. Note that it will create a new folder R_groundhog folder in the working directory (see the message below).
$ docker run --rm -it -v $(pwd):/home/docker \
-w /home/docker -u docker r-base:4.0.2 R
> install.packages("groundhog")
> library(groundhog")
groundhog needs authorization to save files to '/home/docker/R_groundhog'
Enter 'OK' to provide authorization
OK
The groundhog folder path is now:
/home/docker/R_groundhog/groundhog_library/
Loaded 'groundhog' (version:2.1.0) using R-4.0.2
Tips and troubleshooting: https://groundhogR.com
> groundhog.library("
library(ggplot2)",
"2020-10-10")
> library()
# Packages in library ‘/usr/local/lib/R/site-library’:
> packageVersion("ggplot2")
[1] ‘3.3.2’ # the latest version is 3.4.0 on R 4.2.2
> library(ggplot2)
(Current groundhog is v3.1.0) If I don't specify a user in docker run, I'll need to call groundhog.library() twice in order to install packages (eg. "DT" package). In this case I can use groundhog:::save.cookie("copy_instead_of_renaming") before calling groundhog.library().
$ docker run -it --rm rocker/verse:4.3.0 bash
root@58978695ec12:/# R
R version 4.3.0 (2023-04-21) -- "Already Tomorrow"
...
> install.packages("groundhog"); library(groundhog)
> .libPaths()
[1] "/usr/local/lib/R/site-library" "/usr/local/lib/R/library"
> get.groundhog.folder()
[1] "/root/R_groundhog/groundhog_library/"
> groundhog:::save.cookie("copy_instead_of_renaming")
groundhog needs authorization to save files to '/root/R_groundhog/'
Enter 'OK' to provide authorization, and 'NO' not to.
| >OK
Groundhog folder set to: '/root/R_groundhog/groundhog_library/
--- You may change it with`set.groundhog.folder(<path>)`---
Downloading database with information for all CRAN packages ever published
trying URL 'http://s3.wasabisys.com/groundhog/cran.toc.rds'
Content type 'application/octet-stream' length 2010504 bytes (1.9 MB)
==================================================
downloaded 1.9 MB
Downloading database with installation times for all source packages on CRAN
trying URL 'http://s3.wasabisys.com/groundhog/cran.times.rds'
Content type 'application/octet-stream' length 803912 bytes (785 KB)
==================================================
downloaded 785 KB
> groundhog.library("DT", "2023-4-23")
> find.package("DT")
rang
rang: make ancient R code run again
cranly
sessioninfo
tmp = session_info("sessioninfo")
dim(tmp$packages) # [1] 7 11
tmp = session_info("tidyverse")
dim(tmp$packages) # [1] 95 11
Reverse dependence
- http://romainfrancois.blog.free.fr/index.php?post/2011/10/30/Rcpp-reverse-dependency-graph
- Checking reverse dependencies: the tiny way
- pkgcache: Cache 'CRAN'-Like Metadata and R Packages
- How to Assess Usage of your Package
- We can calculate the number of direct, total and indirect dependencies
tinyverse
- https://www.tinyverse.org/
- ggplot2 4.0.0 is coming and why ultimately it’s on YOU to ensure your environments are reproducible
Install packages offline
http://www.mango-solutions.com/wp/2017/05/installing-packages-without-internet/
Install a packages locally and its dependencies
It's impossible to install the dependencies if you want to install a package locally. See Windows-GUI: "Install Packages from local zip files" and dependencies
A minimal R package (for testing purpose)
- https://github.com/joelnitta/minimal. Question: is there a one from CRAN?
- The minimal R package
- Linux command ls -lSr | grep '^-' | head -5
- QuadRoot - the smallest (7002 bytes) R package on CRAN as of 5/10/2023. Others include (starting from the smallest) relen, signibox, freqdist, fdq.
- LifeInsuranceContracts - the smallest (1090 bytes). signibox - the 2nd smallest (1415 bytes) R package on CRAN as of 7/12/2025.
An R package that does not require others during install.packages()
Create a new R package, namespace, documentation
- http://cran.r-project.org/doc/contrib/Leisch-CreatingPackages.pdf (highly recommend)
- https://stat.ethz.ch/pipermail/r-devel/2013-July/066975.html
- Benefit of import in a namespace
- This youtube video from Tyler Rinker teaches how to use RStudio to develop an R package and also use Git to do version control. Very useful!
- Developing R packages by Jeff Leek in Johns Hopkins University.
- R packages book by Hadley Wickham.
- R package primer a minimal tutorial from Karl Broman.
- How to make and share an R package in 3 steps (6/14/2017)
- Package Development tutorial for useR! 2019 Toulouse
- rOpenSci Packages: Development, Maintenance, and Peer Review
- Package development from "Modern R with the tidyverse"
- How to write your own R package (video, 10 minutes)
- Building R packages with devtools and usethis | RStudio (video, thomas mock)
- The Package: learning how to build an R package
- How to write your own R package and publish it on CRAN
- R package development workflow (assuming you’re using macOS or Linux)
- R Package Development Advent Calendar 2025: A Complete Journey 2025 Dec.
Package structure
http://r-pkgs.had.co.nz/package.html. On Linux/macOS, use tree -d DIRNAME to show the directories only. At a minimum, we will have R and man directories.
- ChangeLog
- DESCRIPTION
- MD5
- NAMESPACE
- R/
- zzz.R
- build/
- Package.pdf (eg dplyr)
- vignette.rds
- data/
- demo/
- inst/
- extdata/
- doc/
- FileName.R
- FileName.Rmd
- FileName.html
- include/
- othersYouInclude/
- tinytest/
- CITATION
- man/
- figures/
- src/
- tests/
- testthat
- vignettes/
NAMESPACE and DESCRIPTION
Namespace dependencies not required. If you use import or importFrom in your NAMESPACE file, you should have an entry for that package in the Imports section of your DESCRIPTION file (unless there is a reason that you need to use Depends in which case the package should have an entry in Depends, and not Imports).
license
- What kind of license is the best license for an R package?
- https://cran.r-project.org/web/licenses/
- https://r-pkgs.org/description.html#description-license
- Some Notes on GNU Licenses in R Packages
Install software for PDF output
- Windows: Miktex and pandoc (Create HTML or PDF Files with R, Knitr, MiKTeX, and Pandoc)
- Unix: TeX Live (sudo apt-get install texlive-full) and pandoc
- Mac: MacTex
Windows: Rtools
- (Old) Installing RTools for Compiled Code via Rcpp. Just remember to check the option to include some paths in the PATH environment variable.
- Using Rtools4 on Windows. This version gives more details about the story in Rtools4X versions.
- Testing Rtools43 on R 4.3.3. According to the information on the website: When using R installed by the installer, no further setup is necessary after installing Rtools43 to build R packages from source. I can testify that by running install.packages("jsonlite", type = "source") . We can also verify that make can be found.
Sys.which("make") ## "C:\\rtools40\\usr\\bin\\make.exe"
Screenshots of installation of Rtools44
R CMD
R Installation and Administration
- R CMD build XXX. Note this will not create pdf files for vignettes. The output is a tarball.
- R CMD INSTALL
- R CMD REMOVE
- R CMD SHLIB files. For example, "Rcmd shlib *.f *.c -o surv.dll" on Windows.
- R CMD make
- R CMD check XXX. Useful if we want to create reference manual (PDF file). See R create reference manual with R CMD check.
- R CMD javareconf
usethis package
- https://usethis.r-lib.org/. usethis is a workflow package: it automates repetitive tasks that arise during project setup and development, both for R packages and non-package projects.
- R Package Development 1: Where to Start by John Muschelli. R Package Development playlist.
- create_package(path) with the package name of your choice.
- Sharing with others and helping yourself (slides, use right click menu to save as an HTML file)
- usethis 1.5.0
- usethis 1.6.0
- usethis 2.0.0
Github Actions
To use the Github Actions for continuous integration/CI checks,
- I first follow this to run usethis::use_github_action_check_release(). Once I commit and push the files to Github, Github Actions are kicked off. The R console also tell me to copy and paste a line to add a workflow's badge to README.md.
- Then I modify the yaml file to become this to run a standard check. This took 3m24s to run.
- Then I further delete the 'devel' line (line 23) to reduce one more platform to check. This took 3m to check. My example is on Github (rtoy).
I also try to follow Github Actions with R and create a pkgdown/package documentation web page.
- usethis::use_github_actions("pkgdown") Now go to Github repo's Settings -> Options and scroll down until you see Github Pages. For Source, the page site should be set to being built from the root folder of the gh-pages.
- I have used Jekyll to create a gh-page. I don't need to delete anything for this new gh-pages. I just need to go to the repository setting and (scroll down until we see Github Pages) change the Source of Github Pages to 'gh-pages branch' from 'master branch'.
- The files on the gh-pages branch are generated by Github Actions; these files are not available on my local machine. My location machine only has .github/workflows/pkgdown.yaml file.
Question:
- The workflow file specifies R version and OS platform.
- Right now the workflow file (like pkgdown) is using "r-lib/actions/setup-r@master" that has an "action.yml" file. The r-version is '3.x' only. What about if R 4.0.0 is released?
Packages, webpages and Github
Tutorials by Lisa DeBruine
biocthis
R package depends vs imports
- http://stackoverflow.com/questions/8637993/better-explanation-of-when-to-use-imports-depends
- http://stackoverflow.com/questions/9893791/imports-and-depends
- https://stat.ethz.ch/pipermail/r-devel/2013-August/067082.html
In the namespace era Depends is never really needed. All modern packages have no technical need for Depends anymore. Loosely speaking the only purpose of Depends today is to expose other package's functions to the user without re-exporting them.
load = functions exported in myPkg are available to interested parties as myPkg::foo or via direct imports - essentially this means the package can now be used
attach = the namespace (and thus all exported functions) is attached to the search path - the only effect is that you have now added the exported functions to the global pool of functions - sort of like dumping them in the workspace (for all practical purposes, not technically)
import a function into a package = make sure that this function works in my package regardless of the search path (so I can write fn1 instead of pkg1::fn1 and still know it will come from pkg1 and not someone's workspace or other package that chose the same name)
The distinction is between "loading" and "attaching" a package. Loading it (which would be done if you had MASS::loglm, or imported it) guarantees that the package is initialized and in memory, but doesn't make it visible to the user without the explicit MASS:: prefix. Attaching it first loads it, then modifies the user's search list so the user can see it.
Loading is less intrusive, so it's preferred over attaching. Both library() and require() would attach it.
import() and importFrom()
If our package depends on a package, we need to made some changes. Below we assume the package glmnet in our new package.
- DESCRIPTION: Imports: glmnet
- NAMESPACE: either import(glmnet) to import all functions from glmnet or importFrom(glmnet, cv.glmnet) to import 'cv.glmnet' only
- hello.R: nothing needs to be added
For more resource, see
R package suggests
stringr has suggested htmlwidgets. An error will come out if the suggested packages are not available.
> library(stringr)
> str_view(c("abc", "a.c", "bef"), "a\\.c")
Error in loadNamespace(name) : there is no package called ‘htmlwidgets’
Useful functions for accessing files in packages
- system.file()
- path.package() & find.package(). Note that path.package() requires the package to be loaded.
- file.path(). This is not related to packages.
- normalizePath(). This is not related to packages.
> system.file(package = "batr")
[1] "f:/batr"
> system.file("extdata", "logo.png", package = "cowplot") # Mac
[1] "/Library/Frameworks/R.framework/Versions/4.0/Resources/library/cowplot/extdata/logo.png"
> path.package("batr")
[1] "f:\\batr"
> path.package("ggplot2") # Mac
[1] "/Library/Frameworks/R.framework/Versions/4.0/Resources/library/ggplot2"
# sometimes it returns the forward slash format for some reason; C:/Program Files/R/R-3.4.0/library/batr
# so it is best to add normalizePath().
> normalizePath(path.package("batr"))
> file.path("f:", "git", "surveyor")
[1] "f:/git/surveyor"
Internal functions
RStudio shortcuts
available package
available. Check if a given package name is available to use. It checks the name's validity. Checks if it is used on 'GitHub', 'CRAN' and 'Bioconductor'.
Create an R package
- Your first R package in 1 hour
- Learning To Create an R Package With Deliberate Redundancy - A Note For Myself
Using usethis
New R Package 'foo' -- Updated
- setwd()
- usethis::create_package("foo")
- usethis::use_git(); usethis_github()
- usethis::use_mit_license("Your name")
- usethis::use_r("foo_function")
- usethis::use_package("dplyr") # specify import dependency, e.g. dplyr package
- usethis::use_testthat()
- usethis::use_test("firsttest")
- setwd("..")
- roxygen2::roxygenise()
Using devtools and usethis
- https://www.r-project.org/nosvn/pandoc/devtools.html
- A useful post by Jacob Montgomery. Watch the youtube video there. The process requires 3 components: RStudio software, devtools and roxygen2 (creating documentation from R code) packages.
- MAKING PACKAGES IN R USING DEVTOOLS
- R code workflow from Hadley Wickham.
- Workflow automation tools for package developers
- RStudio:addins part 2 - roxygen documentation formatting made easy
- Inserting a skeleton - Do this by placing your cursor anywhere in the function you want to document and click Code Tools -> Insert Roxygen Skeleton
- Instructions for Creating Your Own R Package. It includes creating a R package with functions written in C++ via Rcpp helper function.
- Creating a simple R package with C++ code to sum “x + 1” by usethis and devtools.
- devtools cheatsheet (2 pages)
- My first R package Part 1, Part 2, Part 3. It uses 3 packages: usethis, roxygen2 and devtools.
- If you love devtools::load_all(), pkgload is where the magic actually happens. pkgload package.
A Typical Streamlined Workflow
- Start a New Package
- Navigate to the directory where you want your package to live.
- Run the following command in your R console:
usethis::create_package("my_package") - This creates the my_package directory with the basic file structure and a .Rproj file.
- Open the Project
- Open the newly created my_package.Rproj file in RStudio. This is the most crucial step as it sets your working directory to the package root.
- Add Your First R Script
- Use this command to create a new R script file:
usethis::use_r("my_function") - This creates the R/my_function.R file and opens it for you.
- Use this command to create a new R script file:
- Write and Document Your Code
- Add your function code to R/my_function.R.
- Use roxygen2 comments to document your function.
- Add a Test File
- Use this command to create a basic test file for your function:
usethis::use_test("my_function") - This creates a test-my_function.R file in the tests/testthat/ directory.
- Use this command to create a basic test file for your function:
- Interactive Development Loop
- Use these commands repeatedly as you work:
devtools::load_all() devtools::test()
- Use these commands repeatedly as you work:
- Generate Documentation
- After writing your roxygen comments, run this command to generate the documentation:
devtools::document()
- After writing your roxygen comments, run this command to generate the documentation:
- Setup the README
- Run this command to create a
README.Rmdfile:usethis::use_readme_rmd()
- Run this command to create a
- Write the README Content
- Open the
README.Rmdfile and add your package description, installation instructions, and examples.
- Open the
- Build the README
- After making changes to the .Rmd file, run this command to generate the README.md file:
devtools::build_readme()
- After making changes to the .Rmd file, run this command to generate the README.md file:
- Perform a Final Check
- Before sharing, run a comprehensive check:
devtools::check() - This will report any issues that need to be fixed.
- Before sharing, run a comprehensive check:
- Version Control and Sharing
- To use Git and GitHub:
usethis::use_git() usethis::use_github() - The rendered README.md will be displayed on your repository's front page.
- To use Git and GitHub:
- Release
- For a release, use one of these commands:
devtools::install_github("your_username/my_package") # share with others devtools::release() # submitting to CRAN
- For a release, use one of these commands:
Using RStudio
- Building, Testing, and Distributing Packages from RStudio.
- Youtube
- How to write your own R package from IDG TECHtalk
- R Package Development playlist by John Muschelli.
- https://youtu.be/Mwe64ziVQXA which does not use RStudio to create a new package
- How to Create and Distribute an R Package.
- The goal of the article to set up an easily installable R package on Github for others to use via remotes::install_github().
- The main tools required are RStudio along with the packages roxygen2 and usethis.
- How to take care of Bioconductor and Github Dependencies
- How to publish your package to Github
- Building a Corporate R Package for Pleasure and Profit
- Toy example*
- In RStudio, click "File" - "New Project" - "New Directory" - "R package". Package name = "a1" (this naming will guarantee this package will be shown on top of all R packages in RStudio). Press 'Create Project'. This new project folder will have necessary files for an R package including DESCRIPTION, NAMESPACE, R/hello.R, man/hello.Rd. The "hello.R" file will be opened in RStudio automatically.
- Create a new R file under a1/R folder. An example of this R file <add.R> containing roxygen comments can be found under here. Pressing Ctrl/Cmd + Shift + D (or running devtools::document()) will generate a man/add.Rd.
- In R, type usethis::use_vignette("my-vignette") to create a new vignette. The new vignette "my-vignette.Rmd" will be saved under "a1/vignettes" subfolder. We can modify the Rmd file as we need.
- In RStudio, click "Build" - "Build Source Package". You will see some messages on the "Build" tab of the top-right panel. Eventually, a tarball "/home/$USERNAME/a1_0.1.0.tar.gz" is created (I create the project under /home/$USERNAME directory).
- We can install the package in R install.packages("~/a1_0.1.0.tar.gz", repos= NULL, type = "source") if we have already created the tarball. Another method is to use RStudio "Build" - "Install and Restart" (Ctrl + Shift + B).
- In RStudio, type help(package = "a1") or click "Packages" tab on the bottom-right panel and click "a1" package. It will show a line "User guides, package vignettes and other documentation". The vignette we just created will be available in HTML, source and R code format.
Using RStudio.cloud
There is a problem. We can use devtools::create("/cloud/project") to create a new package. When it builds the source package, the package file will be located in the root directory of the package. However, if we use a local RStudio to create a source package, the source package will be located in the upper directory.
Binary packages
- No .R files in the R/ directory. There are 3 files that store the parsed functions in an efficient file format. This is the result of loading all the R code and then saving the functions with save().
- A Meta/ directory contains a number of Rds files. These files contain cached metadata about the package, like what topics the help files cover and parsed version of the DESCRIPTION file.
- An html/ directory.
- libs/ directory if you have any code in the src/' directory
- The contents of inst/ are moved to the top-level directory.
Building the tarball
- No matter we uses devtools::build() or the terminal R CMD build MyPkg it will uses run the R code in vignette. Be cautious on the extra time and storage the process incurred.
- If 'cache = TRUE' is used in vignettes, it will create a new subfolder called MyuPkg_cache under the vignettes folder. This takes a lot of space (eg 1GB in some case).
Building the binary
R CMD INSTALL --build MyPkg.tar.gz # OR R CMD INSTALL --build Full_Path_Of_MyPkg
The binary (on Windows) can be installed by install.packages("Mypkg.zip",repos=NULL)
If the installation is successful, it will overwrite any existing installation of the same package. To prevent changes to the present working installation or to provide an install location with write access, create a suitably located directory with write access and use the -l option to build the package in the chosen location.
R CMD INSTALL -l location --build pkg
R folder
- https://r-pkgs.org/r.html
- zzz.R. .onLoad() function.
- R: How to run some code on load of package?
See an example from DuoClustering2018.
#' @importFrom utils read.csv
.onLoad <- function(libname, pkgname) {
fl <- system.file("extdata", "metadata.csv", package = "DuoClustering2018")
titles <- utils::read.csv(fl, stringsAsFactors = FALSE)$Title
ExperimentHub::createHubAccessors(pkgname, titles)
}
Note that the environment of a function from the DuoClustering2018 package is not the package name.
environment(clustering_summary_filteredExpr10_TrapnellTCC_v2) <environment: 0x7fe01dbe7dd0>
Q: where is the definition of DuoClustering2018::clustering_summary_filteredExpr10_TrapnellTCC_v2()? A: createHubAccessors() - Creating A Hub Package: ExperimentHub or AnnotationHub.
data
http://r-pkgs.had.co.nz/data.html
Three ways to include data in your package.
- If you want to store binary data and make it available to the user, put it in data/. This is the best place to put example datasets.
- If you want to store parsed data, but not make it available to the user, put it in R/sysdata.rda. This is the best place to put data that your functions need.
- If you want to store raw data, put it in inst/extdata. See External data. An example from tximportData package.
# grep -r extdata /home/brb/R/x86_64-pc-linux-gnu-library/4.0 logo_file <- system.file("extdata", "logo.png", package = "cowplot") orgDBLoc = system.file("extdata", "org.Hs.eg.sqlite", package="org.Hs.eg.db") # grep -r readRDS /home/brb/R/x86_64-pc-linux-gnu-library/4.0 patient.data <- readRDS("assets/coxnet.RDS")
How to distribute data with your R package
Rd file
Vignette
- How to Create Vignettes That Really Pull a Room Together
- See examples from packages like magrittr (Rmd, jpg) or tidyr (Rmd, csv) or rvest (Rmd, png) or DESeq2.
- Many vignette uses output: rmarkdown::html_vignette which will expand the width to the page. This one uses output: BiocStyle::html_document which will have a narrow width.
- Add a static pdf vignette to an R package
- devtools::build_vignettes(). The files are copied in the 'doc' directory and an vignette index is created in 'Meta/vignette.rds', as they would be in a built package.
- build_vignette can't find functions
Long execution for R code in vignette
- Caching chunks in RStudio does not work
- caching doesn't seem to work here (i.e. when used in a document that is a vignette)
- How to Add a Vignette to a Package in RStudio
- Optimal workflows for package vignettes
NEWS
Why and how maintain a NEWS file for your R package?
README.Rmd & README.md files
See Releasing a package from R packages by Hadley Wickham.
How to convert .Rmd into .md in R studio?
Example: ggplot2 repository at Github
It seems RStudio cannot create TOC for *.md files. glmnet package creates TOC of its vignette by itself. Visual Studio Code has an extension to do that.
badge
badger: Badge for R Package
tests folder and testthat package
- testthat package.
- testthat: Get Started with Testing 2011
- Automated testing with 'testthat' in practice
- Lots of packages suggest testthat; for example princurve, glmnetUtils.
- How to run a package's testthat tests
- Building, Testing, and Distributing Packages from RStudio
- Run Tests button. See RStudio release notes from RStudio v1.2 - April 8, 2019.
- Working with the RStudio IDE
- Unit Tests in R 2019
- The result of dput can be used in expect_equal().
- A small gotcha when comparing lists using testthat
.Rbuildignore
Non-standard files/directories, Rbuildignore and inst
URL checker
What is a library?
A library is simply a directory containing installed packages.
You can use .libPaths() to see which libraries are currently active.
.libPaths() lapply(.libPaths(), dir)
Object names
- Variable and function names should be lower case.
- Use an underscore (_) to separate words within a name (reserve . for S3 methods).
- Camel case is a legitimate alternative, but be consistent! For example, preProcess(), twoClassData, createDataPartition(), trainingRows, trainPredictors, testPredictors, trainClasses, testClasses have been used in Applied Predictive Modeling by Kuhn & Johnson.
- Generally, variable names should be nouns and function names should be verb.
Spacing
- Add a space around the operators +, -, \ and *.
- Include a space around the assignment operators, <- and =.
- Add a space around any comparison operators such as == and <.
Indentation
- Use two spaces to indent code.
- Never mix tabs and spaces.
- RStudio can automatically convert the tab character to spaces (see Tools -> Global options -> Code).
\dontrun{}
- Writing R Extensions > Documenting functions and search for dontrun
- How to not run an example using roxygen2?
- What does "not run" mean in R help pages? See ?example .
formatR and lintr package
Use formatR package to clean up poorly formatted code
install.packages("formatR")
formatR::tidy_dir("R")
Another way is to use the lintr package (lint).
install.packages("lintr")
lintr:::lin_package()
Rcpp
Thirteen Simple Steps for Creating An R Package with an External C++ Library
C library
Using R — Packaging a C library in 15 minutes
Data package
Chapter 12 Create a data package from rstudio4edu
Minimal R package for submission
https://stat.ethz.ch/pipermail/r-devel/2013-August/067257.html and CRAN Repository Policy.
Create R Windows Binary on non-Windows OS
- Create R Windows Binary from .tar.gz linux
- win-builder from r-project.org
r-hub/rhub package: the R package builder service
https://github.com/r-hub/proposal, R-hub v2 2024/4.
- rhub 1.1.1 is on CRAN! 2019/4/8
- R package developers, why should you care about R-hub? 2019/3/26
- Local Linux checks with Docker
- https://www.rstudio.com/resources/videos/r-hub-overview/
- http://blog.revolutionanalytics.com/2016/10/r-hub-public-beta.html
- Setting up continuous multi-platform R package building, checking and testing with R-Hub, Docker and GitLab CI/CD for free, with a working example 2019/4/27
- How to keep up with CRAN policies and processes? 2019/5/29
- R-hub usage in a few figures
# Today 1/1/2020 $ git clone https://github.com/arraytools/rtoy.git $ rm -rf rtoy/.git $ rm rtoy/.gitignore rtoy/_config.yml $ R > install.packages("devtools", repos = "https://cran.rstudio.com") > install.packages("rhub") > devtools::install_github("r-hub/sysreqs") #needed before calling local_check_linux() > pkg_path <- "~/Downloads/rtoy" > chk <- local_check_linux(pkg_path, image = "rhub/debian-gcc-release") ─ Building package Container name: 6dca434d-84f9-42e2-ab83-b8c364594476-2 It will _not_ be removed after the check. R-hub Linux builder script v0.10.0 (c) R Consortium, 2018-2019 Package: /tmp/RtmpviOCT1/file470972a3c4ab/rtoy_0.1.0.tar.gz Docker image: rhub/debian-gcc-release Env vars: R CMD check arguments: Unable to find image 'rhub/debian-gcc-release:latest' locally latest: Pulling from rhub/debian-gcc-release Digest: sha256:a9e01ca57bfd44f20eb6719f0bfecdd8cf0f59610984342598a53f11555b515d Status: Downloaded newer image for rhub/debian-gcc-release:latest Sysreqs platform: linux-x86_64-debian-gcc No system requirements >>>>>==================== Installing system requirements 8fa4f66e41954b4ed1112eb72e76683c28345863fcc7e260edac6a9a30387fed >>>>>==================== Starting Docker container 2dfae75c9de95b598ff8d9cd19d3cfc7ffe7f16edbc3d4f81c896e4e6b956ebd ls: cannot access '/opt/R-*': No such file or directory > source('https://bioconductor.org/biocLite.R') Error: With R version 3.5 or greater, install Bioconductor packages using BiocManager; see https://bioconductor.org/install Execution halted Error in run(bash, c(file.path(wd, "rhub-linux.sh"), args), echo = TRUE, : System command error
Moreover, it create a new Docker image and a new Docker container. We need to manually clean them:-(
Running a check on its own (remote) server works
> check_on_linux("rtoy") # will run remotely.
# We need to verify the email and enter a token.
# We will get a report and a full build log.
# The report includes both the linux command and the log from the server.
Now I go back to the original method.
$ R
> install.packages("tinytex")
> tinytex::install_tinytex()
> q()
$ exit
$ sudo apt-get install texinfo
$ R CMD build rtoy
$ R CMD check rtoy_0.1.0.tar.gz
# Install pandoc
$ wget https://github.com/jgm/pandoc/releases/download/2.9.1/pandoc-2.9.1-1-amd64.deb
$ sudo dpkg -i pandoc-2.9.1-1-amd64.deb
$ R CMD check --as-cran rtoy_0.1.0.tar.gz
# Ignore a 'Note' https://stackoverflow.com/a/23831508
rcmdcheck
Run R CMD check from R and Capture Results
The rcmdcheck package was used by Github Actions for R language from r-lib/R infrastructure.
CRAN check API
- Overview of the CRAN checks API 2019/6/10
- update 2019/10/9
Continuous Integration
Travis-CI (Linux, Mac)
- A Beginner's Guide to Travis-CI
- http://johnmuschelli.com/neuroc/getting_ready_for_submission/index.html#61_travis
Continuous Integration: Appveyor (Windows)
- Appveyor is a continuous integration service that builds projects on Windows machines.
- http://johnmuschelli.com/neuroc/getting_ready_for_submission/index.html#62_appveyor
Github Actions
- See usethis package.
- Automating R package checks across platforms with GitHub Actions and Docker in a portable way. No need to use "usethis" package.
precommit
Submit packages to cran
- https://cran.r-project.org/submit.html
- http://f.briatte.org/r/submitting-packages-to-cran
- Submitting your first package to CRAN, my experience
- Get your R package on CRAN in 10 steps June 2018
- Preparing Your Package for for Submission by John Muschelli
- https://builder.r-hub.io/. See here.
- The most annoying warning for CRAN submission
- How to write your own R package and publish it on CRAN devtools::release(), July 2020
- A NOTE on URL checks of your R package
- Releasing a package from the book "R packages" by Hadley Wickham.
- I got my first package onto CRAN, and YOU CAN TOO
- Submitting vroom to CRAN, LIVE!
Windows
Everything you should know about WinBuilder
Other tips/advice
- Top 10 tips to make your R package even more awesome
- GoodPractice Advice on R Package Building.
C/Fortran
Rmath.h
For example pnorm5() was used by survS.h by survival package (old version) .
Makevars
- Writing R Extensions
- The order of inclusion of makefiles for a package which does not have a src/Makefile file is (Section 1.1.5 Package subdirectories) on a Unix-alike system
- src/Makevars
- R_HOME/etc/R_ARCH/Makeconf
- R_MAKEVARS_SITE, R_HOME/etc/R_ARCH/Makevars.site
- R_HOME/share/make/shlib.mk
- R_MAKEVARS_USER, ~/.R/Makevars-platform, ~/.R/Makevars
- The order of inclusion of makefiles for a package which does not have a src/Makefile file is (Section 1.1.5 Package subdirectories) on a Unix-alike system
- R Installation and Administration
- Section 6.3.3 Customizing package compilation: This section explains that users can create a personal Makevars file to set their own compiler flags (like CC, CFLAGS, CXXFLAGS, etc.).
- Platform Specifics: It notes that on Unix-alikes (including macOS and Linux), the file is located at ~/.R/Makevars. On Windows, it is typically found at Documents/.R/Makevars.win
- Section 1.2.1 (Using Makevars): It describes how Makevars files (both inside a package's src/ directory and in the user's home directory) are used to define macros for the make utility.
- Variable Precedence: It explains that user-level settings in ~/.R/Makevars will generally override the defaults set when R was configured, allowing you to use different compilers or include extra library paths (like the gettext path in the benchmarKIN package case.
Packages includes Fortran
Some useful packages containing fortran code.
- survC1
- quantreg
- nlme
- glmnet
- bsamGP (FORTRAN 90). This is searched by using https://www.r-pkg.org/. I am not able to get what I need using the website.
My experience on M1 macOS
- (Outdated) On mac, gfortran (6.1) (X86 not arm) can be downloaded from CRAN. It will be installed onto /usr/local/gfortran. This can be confirmed by:
mkdir temp cd temp xar -xf ../gfortran-6.1.pkg lsbom Bom
Note that the binary will not be present in PATH. So we need to run the following command to make gfortran availalble. <spre> sudo ln -s /usr/local/gfortran/bin/gfortran /usr/local/bin/gfortran
- Tested in R 4.4.1, R is looking for /opt/gfortran/bin/gfortran.
- Installing gfortran on MacBook with Apple M1 chip for use in R.
- See https://mac.r-project.org/tools/. Don't follow https://cran.r-project.org/bin/macosx/tools/ to download gfortran.
- R-universe now builds MacOS ARM64 binaries for use on Apple Silicon (aka M1/M2/M3) systems.
- Instruction:
- Download "gfortran-12.2-universal.pkg" from https://mac.r-project.org/tools/,
- Double click the pkg file
- gfortran will be installed on "/opt/gfortran". When R installed a package containing FORTRAN code, it will automatically look for gfortran from /opt/gfortran/bin/gfortran. So it is not necessary to make gfortran available in $PATH.
- Note I already had gfortran installed via homebrew for some software. But it is not used by R.
$ brew uninstall gfortran Error: Refusing to uninstall /opt/homebrew/Cellar/gcc/14.1.0_1 because it is required by gstreamer, numpy, openblas and openvino, which are currently installed. You can override this and force removal with: brew uninstall --ignore-dependencies gfortran $ which -a gfortran /opt/homebrew/bin/gfortran
- In order to change mess up anything, it is better to call export PATH="/usr/local/bin:$PATH" when we want to install R packages containing fortran code.
- A useful tool to find R packages containing Fortran code is pkgsearch package. Note
- The result is not a comprehensive list of packages containing Fortran code.
- It seems the result is the same as I got from https://www.r-pkg.org
> pkg_search("Fortran") - "Fortran" ------------------------------------ 69 packages in 0.009 seconds - # package version by @ title 1 100 covr 3.3.2 Jim Hester 12d Test Coverage for Packages 2 91 inline 0.3.15 Dirk Eddelbuettel 1y Functions to Inline C, ... 3 43 randomForest 4.6.14 Andy Liaw 2y Breiman and Cutler's Ra... 4 39 deSolve 1.24 Thomas Petzoldt 4M Solvers for Initial Val... 5 27 mnormt 1.5.5 Adelchi Azzalini 3y The Multivariate Normal... 6 26 minqa 1.2.4 Katharine M. Mullen 5y Derivative-free optimiz... 7 24 rgcvpack 0.1.4 Xianhong Xie 6y R Interface for GCVPACK... 8 22 leaps 3.0 Thomas Lumley 3y Regression Subset Selec... 9 21 akima 0.6.2 Albrecht Gebhardt 3y Interpolation of Irregu... 10 20 rootSolve 1.7 Karline Soetaert 3y Nonlinear Root Finding,... > more() - "Fortran" ------------------------------------ 69 packages in 0.009 seconds - # package version by @ title 11 15 BB 2019.10.1 Paul Gilbert 11d Solving and Optimizing L... 12 15 limSolve 1.5.5.3 Karline Soetaert 2y Solving Linear Inverse M... 13 14 insideRODE 2.0 YUZHUO PAN 7y insideRODE includes buil... 14 13 earth 5.1.1 Stephen Milborrow 7M Multivariate Adaptive Re... 15 13 cluster 2.1.0 Martin Maechler 4M "Finding Groups in Data"... 16 13 spam 2.3.0.2 ORPHANED 11h SPArse Matrix 17 12 diptest 0.75.7 Martin Maechler 4y Hartigan's Dip Test Stat... 18 10 pbivnorm 0.6.0 Brenton Kenkel 5y Vectorized Bivariate Nor... 19 7 optmatch 0.9.12 Mark M. Fredrickson 17d Functions for Optimal Ma... 20 7 lsei 1.2.0 Yong Wang 2y Solving Least Squares or... - Another way to find out packages containing fortran code is to run rsync to download src/contrib directory from CRAN and then use grep to find these packages. Note: the source packages takes about 8GB space (2019-10-28).
mkdir ~/Downloads/cran rsync -avz --delete cran.r-project.org::CRAN/src/contrib/*.tar.gz ~/Downloads/cran/ rsync -avz --delete cran.r-project.org::CRAN/src/contrib/PACKAGES ~/Downloads/cran/ rsync -avz --delete cran.r-project.org::CRAN/src/contrib/PACKAGES.gz ~/Downloads/cran/ cd ~/Downloads/cran find . -xtype l -delete # remove broken symbolic links touch tmp for f in *.gz; do tar -tzvf $f | grep -E "(\.f|\.f90|\.f95)$" |& tee -a tmp done
to check if the tarball contains Fortran 77/90 code.
Now to see all packages names we can process it in R
library(magrittr) library(stringr) x <- read.table("~/Downloads/tmp", stringsAsFactors = F) strsplit(x$V6, "/") %>% sapply(function(x) x[1]) %>% unique() # 415 packages strsplit(x$V6, "/") %>% sapply(function(x) x[1]) %>% table() %>% sort() # how many Fortran files in each package str_subset(x$V6, "\\.f90$") %>% strsplit("/") %>% sapply(function(x) x[1]) %>% unique() # Fortran 90 only packages str_subset(x$V6, "\\.f95$") %>% strsplit("/") %>% sapply(function(x) x[1]) %>% unique() # Fortran 95 only packages str_subset(x$V6, "BayesFM") # f95
Misc
Datasets in R packages
https://vincentarelbundock.github.io/Rdatasets/datasets.html
Turn your analysis into a package
Build R package faster using multicore
http://www.rexamine.com/2015/07/speeding-up-r-package-installation-process/
The idea is edit the /lib64/R/etc/Renviron file (where /lib64/R/etc/ is the result to a call to the R.home() function in R) and set:
MAKE='make -j 8' # submit 8 jobs at once
Then build R package as regular, for example,
$ time R CMD INSTALL ~/R/stringi --preclean --configure-args='--disable-pkg-config'
suppressPackageStartupMessages() and .onAttach()
KernSmooth package example.
It is Time for CRAN to Ban Package Ads
suppressPackageStartupMessages(library("dplyr"))
fusen package
- {fusen} is now available on CRAN!
- (Videos) How To Build A Package With The "Rmd First" Method, fusen::inflate()
Identifying R Functions & Packages Used in GitHub Repos
Identifying R Functions & Packages Used in GitHub Repos: funspotr package
CRANalerts
R-universe
- How to create your personal CRAN-like repository on R-universe
- Downloading snapshots and creating stable R packages repositories using r-universe