ScRNA: Difference between revisions
| Line 1,323: | Line 1,323: | ||
* We need to install [https://sourceforge.net/projects/mcmc-jags/files/JAGS/4.x/Mac%20OS%20X/ jags] software before we install the required package '''rjags'''. It seems OK to install this old version 4.3.0 of software on macOS Catalina 10.15.7. To install jags, right click the file 'JAGS-4.3.0.mpkg' and choose 'open'. Then follow the instruction of the installer to complete the installation. | * We need to install [https://sourceforge.net/projects/mcmc-jags/files/JAGS/4.x/Mac%20OS%20X/ jags] software before we install the required package '''rjags'''. It seems OK to install this old version 4.3.0 of software on macOS Catalina 10.15.7. To install jags, right click the file 'JAGS-4.3.0.mpkg' and choose 'open'. Then follow the instruction of the installer to complete the installation. | ||
* https://www.jianshu.com/c/4c982303e19f?order_by=top | * https://www.jianshu.com/c/4c982303e19f?order_by=top | ||
** It seems the goal is to use the CNV data to cluster cells... All cells that clustered together with spiked in controls were labeled "nontumor" whereas the remaining clusters were labels as 'tumor'. | |||
** [https://mp.weixin.qq.com/s?__biz=MzI1Njk4ODE0MQ==&mid=2247485693&idx=1&sn=57ffc4d2cf4a6a0b7ddd24d308baa1c2&scene=21#wechat_redirect 使用inferCNV分析单细胞转录组中拷贝数变异]. Or follow the help of plot_cnv(), infercnv::run(). | ** [https://mp.weixin.qq.com/s?__biz=MzI1Njk4ODE0MQ==&mid=2247485693&idx=1&sn=57ffc4d2cf4a6a0b7ddd24d308baa1c2&scene=21#wechat_redirect 使用inferCNV分析单细胞转录组中拷贝数变异]. Or follow the help of plot_cnv(), infercnv::run(). | ||
** [https://www.jianshu.com/p/2ab27d11be23 CNS图表复现13—使用inferCNV来区分肿瘤细胞的恶性与否]. 因为10X技术出来的单个细胞的reads数量太少,检测到的基因数量太少,但是inferCNV更新后有一个参数是cutoff ,就很清楚的指出来了:cutoff=1 works well for Smart-seq2, and cutoff=0.1 works well for 10x Genomics 说明它应用于10X单细胞转录组数据是没有问题的。 | ** [https://www.jianshu.com/p/2ab27d11be23 CNS图表复现13—使用inferCNV来区分肿瘤细胞的恶性与否]. 因为10X技术出来的单个细胞的reads数量太少,检测到的基因数量太少,但是inferCNV更新后有一个参数是cutoff ,就很清楚的指出来了:cutoff=1 works well for Smart-seq2, and cutoff=0.1 works well for 10x Genomics 说明它应用于10X单细胞转录组数据是没有问题的。 | ||
Revision as of 18:39, 4 October 2021
Resource
- https://github.com/seandavi/awesome-single-cell#tutorials-and-workflows List of software packages for single-cell data analysis collected by Sean Davis
- Single-Cell RNA-Sequencing: Assessment of Differential Expression Analysis Methods by Dal Molin et al 2017.
- Normalization and noise reduction for single cell RNA-seq experiments by Bo Ding et al 2015.
- A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor by Lun 2016.
- Gene length and detection bias in single cell RNA sequencing protocols and the script & data are available online. Belinda Phipson1 et al 2017.
- Bioconductor workflow for single-cell RNA sequencing: Normalization, dimensionality reduction, clustering, and lineage inference in ISCB (International Society for Computational Biology Community Journal). It has open peer reviews too.
- scRNA-seq analysis workflow by Roman Hillje
- Welcome to the Tidyverse from the Journal of Open Source Software. It has open peer reviews too.
- Missing data and technical variability in single-cell RNA-sequencing experiments
- A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications 2017
- How to design a single-cell RNA-sequencing experiment: pitfalls, challenges and perspectives by Alessandra Dal Molin 2018
- Crafting a blueprint for single-cell RNA sequencing 2021
- Current best practices in single‐cell RNA‐seq analysis: a tutorial 2019
- DSAVE: Detection of misclassified cells in single-cell RNA-Seq data.
- Top Benefits of Using the Technique of Single Cell RNA-Seq
- Systematic comparison of high-throughput single-cell RNA-seq methods for immune cell profiling 2021
- The triumphs and limitations of computational methods for scRNA-seq Kharchenko 2021
- Single-cell RNA-seq data analysis workshop from Bioinformatics Training at the Harvard Chan Bioinformatics Core
- https://www.plob.org/article/21723.html 单细胞RNA测序 中文網站
- Novartis/scRNAseq_workflow_benchmark
- Top Benefits of Using the Technique of Single Cell RNA-Seq.
- Gain Better Understanding cell types
- Analysis of Rare Cell Types
- Reveals Hidden Differences
- Detection of Cancer Stem Cells
- Enables exploring the Complex Systems
- Describes the Regulatory Networks and Developmental Trajectories
- Videos
- Analysis of single cell RNA-seq data - Lecture 1 by BioinformaticsTraining
- Single-cell isolation by a modular single-cell pipette for RNA-sequencing by labonachipVideos
- Single Cell RNA Sequencing Data Analysis (youtube)
- Introduction to Bioinformatics and Computational Biology Xiaole Shirley Liu (ebook format)
- ebooks
- Analysis of single cell RNA-seq data Ruqian Lyu, PuXue Qiao, and Davis J. McCarthy
- Analysis of single cell RNA-seq data (ebook, Kiselev et al 2019, University of Cambridge Bioinformatics training unit, SingleCellExperiment & Seurat) and the paper Tutorial: guidelines for the computational analysis of single-cell RNA sequencing data Andrews 2020. A docker image is available.
- ANALYSIS OF SINGLE CELL RNA-SEQ DATA (ebook) Ashenberg et al 2019 from Broad institute. Seurat package was used.
- Analysis of single cell RNA-seq data (ebook) Lyu et al. 2019. Seurate and SingleCellExperiment.
Workshops
Library preparation
- Smart-Seq2
- Drop-Seq
- Fluidigm-C1
Pipeline
- scFlow: A Scalable and Reproducible Analysis Pipeline for Single-Cell RNA Sequencing Data. https://github.com/combiz/scFlow
Public data
- https://www.biostars.org/p/208973/
- Collection of Public Single-Cell RNA-Seq Datasets
- Single cell portal https://singlecell.broadinstitute.org/single_cell
- https://cells.ucsc.edu/
- https://www.humancellatlas.org/
- Tung dataset, data.
- SRA/GSE
- cellranger count help
- SRR7611046 and SRR7611048 from Sequence Read Archive (SRA) pages
- SRP073767 PBMC data. GSE29087 Mouse Embryonic Data. GSE64016 Human Embryonic Data. See Normalization Methods on Single-Cell RNA-seq Data: An Empirical Survey Lytal 2020
- GSE92332 from Scripts for "Current best-practices in single-cell RNA-seq: a tutorial"
- BP4RNAseq package. SRR11402955, SRR11402974. Note several dependent tools need to be installed manually.
- GSE75790. See Comparative Analysis of Single-Cell RNA Sequencing Methods Ziegenhain 2017
- GSE137804/SRP222837. 22 samples (GSM or SRX numbers), 42 runs (SRR numbers). The fastq.gz files are also directly available in European Nucleotide Archive where we can see 14 cells have only Read 2 data without Read 1 data.
- GSE29087, GSE94820, GSE67835, GSE65528, GSE70850 from POWSC - Simulation, power evaluation and sample size recommendation for single-cell RNA-seq, Su 2020.
- GSE81076, GSE86473, GSE85241, E-MTAB-5061 from the mutual nearest neighbors (MNNs) method Haghverdi 2018
scRNAseq package
scRNAseq package from Bioconductor.
ExperimentHub package used by scRNAseq, DuoClustering2018 and other packages.
SRAToolkit
- https://hpc.nih.gov/apps/sratoolkit.html. source code, Download SRA sequences from Entrez search results
- Downloading paired end fastq from SRA, fasterq-dump only returns one file for a paired-end sample for scRNA-seq data. The solution fasterq-dump + '--include-technical -S' option according to https://github.com/ncbi/sra-tools/issues/399 (SRR9169172 is scRNA-Seq case) works. It yields the same number of reads when I check using the wc -l command though the md5sum are different.
- fasterqDump() from the geomedb package.
- SRA Toolkit: using fastq-dump vs fasterq-dump - discrepancies in output?
- SRR2048331.
- SRR12148213 (from SRP222837). Note the corresponding sample has 8 runs.
sinteractive --gres=lscratch:800 --cpus-per-task=6 --mem=40g
# the required memory can be checked by using the '''jobload''' command
# the required local disk space can be obtained by 'ssh cnXXX' and run 'du -sh /lscratch/JobID'
# where the JobID is obtained through the jobload command
module load sratoolkit
fasterq-dump -t /lscratch/$SLURM_JOBID SRR2048331 # 1 min, 3.3G fastq file
fastq-dump --split-3 SRR2048331 # 2 min 30 sec
pigz -p6 SRR12148211*.fastq # vs gzip. 6 threads are used here
# do not need to split threads,
# do not need to run separately
fastq-dump --split-3 --gzip SRR2048331 # 658M SRR2048331.fastq.gz (vs 626.9Mb in sra format)
fastq-dump --split-3 --gzip SRR12148213 # 862M + 2.5G for fastq.gz files (vs 2Gb in sra format)
# 6.5G + 15G for fastq files
# fastq/fastq.gz = 15/2.5 = 6
# fastq.gz/sr = 3.4/2 = 1.7
# This is equivalent to
# prefetch SRR12148213; cd SRR12148213
# fastq-dump --split-3 --gzip SRR12148213.sra # 45 min
# The 'prefetch' will create a new folder SRR12148213 and download SRR12148213.sra into it.
# Note the file sizes from _1.fastq and _2.fastq are quite different.
# Another run. Check number of reads.
$ zcat SRR12148214_1.fastq.gz | wc -l
150635300
$ zcat SRR12148214_2.fastq.gz | wc -l
150635300
fasterq-dump -t /lscratch/$SLURM_JOB_ID \
--split-files \
-O /lscratch/$SLURM_JOBID SRR13526458; \
pigz -p6 /lscratch/$SLURM_JOBID/SRR13526458*.fastq; \
cp /lscratch/$SLURM_JOBID/SRR13526458*.fastq.gz ~/PDACPDX
sbatch --gres=lscratch:800 --mem=40g --cpus-per-task=6 --time=12:00:00 myscript
And the fasterq-dump's wiki and the help page
$ fasterq-dump --help
Usage: fasterq-dump [ options ] [ accessions(s)... ]
Parameters:
accessions(s) list of accessions to process
Options:
-o|--outfile <path> full path of outputfile (overrides usage
of current directory and given accession)
-O|--outdir <path> path for outputfile (overrides usage of
current directory, but uses given
accession)
-b|--bufsize <size> size of file-buffer (dflt=1MB, takes
number or number and unit)
-c|--curcache <size> size of cursor-cache (dflt=10MB, takes
number or number and unit)
-m|--mem <size> memory limit for sorting (dflt=100MB,
takes number or number and unit)
-t|--temp <path> path to directory for temp. files
(dflt=current dir.)
-e|--threads <count> how many threads to use (dflt=6)
-p|--progress show progress (not possible if stdout used)
-x|--details print details of all options selected
-s|--split-spot split spots into reads
-S|--split-files write reads into different files
-3|--split-3 writes single reads into special file
--concatenate-reads writes whole spots into one file
-Z|--stdout print output to stdout
-f|--force force overwrite of existing file(s)
-N|--rowid-as-name use rowid as name (avoids using the name
column)
--skip-technical skip technical reads
--include-technical explicitly include technical reads
-P|--print-read-nr include read-number in defline
-M|--min-read-len <count> filter by sequence-lenght
--table <name> which seq-table to use in case of pacbio
--strict terminate on invalid read
-B|--bases <bases> filter output by matching against given
bases
-A|--append append to output-file, instead of
overwriting it
--ngc <path> <path> to ngc file
--perm <path> <path> to permission file
--location <location> location in cloud
--cart <path> <path> to cart file
--disable-multithreading disable multithreading
-V|--version Display the version of the program
-v|--verbose Increase the verbosity of the program
status messages. Use multiple times for
more verbosity.
-L|--log-level <level> Logging level as number or enum string.
One of
(fatal|sys|int|err|warn|info|debug) or
(0-6) Current/default is warn
--option-file file Read more options and parameters from the
file.
-h|--help print this message
"fasterq-dump" version 2.10.9
Note that fastq.gz vs fastq file size is about 1:8 ratio.
$ ls -lhogt | sed 's/^[^ ][^ ]* *[^ ][^ ]* //' 23G May 7 11:35 SRR10156301_2.fastq 18G May 7 11:35 SRR10156301_1.fastq 3.1G May 7 11:02 SRR10156301_2.fastq.gz 1.4G May 7 11:02 SRR10156301_1.fastq.gz
Simulation
A benchmark study of simulation methods for single-cell RNA sequencing data, Blog
Splatter: Simulation Of Single-Cell RNA Sequencing Data
biorxiv. Genome Biol. 2017, splatter package. We can simulate data using 1) estimated parameters from an existing SingleCellExperiment object, or 2) user's specified parameters.
suppressPackageStartupMessages({
library(splatter)
library(scater)
})
library(scRNAseq)
sce2 <- MacoskoRetinaData()
sce2
ncells <- 1000
params <- splatEstimate(as.matrix(assays(sce2)$counts[, 1:ncells]))
sim <- splatSimulate(params)
comparison <- compareSCEs(list(MacoskoRetina = sce2[, 1:ncells], Splat = sim))
ggplot(comparison$ColData, aes(x = sum, y = detected, colour = Dataset)) +
geom_point()
Preprocess
What does single-cell RNA sequencing look like? Ideal droplet containing both a cell and RNA-capture reagents.
fastq file format, UMI
- Single-cell RNA-seq data - raw data to count matrix
- Reads with different UMI are biological duplicates - each read should be counted.
- Reads with the same UMI are are technical duplicates - the UMIs should be collapsed to be counted as a single read.
- Read1 contains barcode (represents cells) and UMI (represents reads), Read2 contains transcript sequence.
- One case from scruff vignette.
- Extract UMI from fastq using the UMI-tools and Extracting UMI sequences from paired-end reads.
- *How 10X RNA-Seq Works. Cell level barcode (16bp) – same for all RNAs in a cell. UMI (10bp) – unique for one RNA in one cell.
- Theory Refresher and Software Overview: Cell Ranger by ELIXIR-IIB Training Platform. More details on Reads 1 & 2.
- 提取barcode和过滤reads, umi_tools for dealing with UMIs and cell barcodes (簡書). 10X is different from Drop-seq.
umi_tools extract --bc-pattern=CCCCCCCCCCCCCCCCNNNNNNNNNN \ --stdin hgmm_100_R1.fastq.gz \ --stdout hgmm_100_R1_extracted.fastq.gz \ --read2-in hgmm_100_R2.fastq.gz \ --read2-out=hgmm_100_R2_extracted.fastq.gz \ --filter-cell-barcode \ --whitelist=whitelist.txt - Barcode extraction for Drop-seq. For Drop-seq, the current protocol includes a 12bp CB, followed by an 8bp UMI.
--extract-method=string --bc-pattern=CCCCCCCCCCCCNNNNNNNN
- *Pre-processing of 10X Single-Cell RNA Datasets. 10x Chromiumv2 and Chromiumv3 Chemistries/reagent kits have the same 16bp of CB but different bp of UMI.
- GSM4654672 also confirms Read 1 & Read 2 contents.
- UMI (Unique molecular identifier)
- Wikipedia
- Single cell tutorial from the documentation of UMI-tools (Tools for dealing with Unique Molecular Identifiers).
- What are UMIs and why are they used in high-throughput sequencing? UMI sequence information in conjunction with alignment coordinates enables grouping of sequencing data into read families representing individual sample DNA or RNA fragments. UMIs are especially helpful for very low input or high depth sequencing.
- UMI-count modeling and differential expression analysis for single-cell RNA sequencing. read-count VS UMI-count. Data & code are available.
- Unique Molecular Identifiers – the problem, the solution and the proof
- CellRanger count
Cell ranger
- Chromium Single Cell Gene Expression
- Gene Expression Algorithms Overview
- Prebuild reference genomes/packages
- You can process between 1–8 samples in one run, loading up to 10,000 cells per sample and other statistics
- You can sequence up to 25,000 cells in each lanes of the chip and this chip has 8 lanes on there. So potentially you could process 200,000 cells.
- Note that for the Single Cell Gene Expression assay, it is possible to target up to 30,000 cells per library if using the 3' CellPlex Kit for Cell Multiplexing. Cell multiplets can be bioinformatically identified and filtered out when using the 3' CellPlex Kit.
- What is the recommended sequencing depth for Single Cell 3' and 5' Gene Expression libraries?
- 10X Genomics Chromium Single-Cell System
- Installing cell ranger - 10X Genomics
- System Requirements. How CPU and Memory Affect Runtime.
- Running cellranger count,
- Understanding Output & cellranger count Web Summary
- The Cell Ranger User Interface. By default, this UI is exposed at an operating-system assigned port, with a randomly-generated authentication token to restrict access.
- *Specifying Input FASTQ Files for 10x Pipelines,
- SRA上那些奇怪的10x Genomics数据 如何解決有些 RUN 只有一个 fastq文件
- How do I prepare Sequence Read Archive (SRA) data from NCBI for Cell Ranger?
- What is the difference between --split-files and --split-3 when using fastq-dump They are the same when I use diff or cmp or md5sum to compare SRR12148212_1.fastq file (7.6G). The files are downloaded by fasterq-dump without or with the --split-files option. Note using md5sum to compare the compressed (gz) files may shows differences though the file sizes are the same. So we should be careful when we use md5sum to compare binary files.
- pbmc3k. The data is saved in filtered_gene_bc_matrices/hg19 directory with 3 files, barcodes.tsv (45K), genes.tsv (798K) and matrix.mtx (27M).
- Getting started with Cell Ranger by Dave Tang
- Workshop from UC Davis
- *10x genomics single-cell RNAseq analysis from SRA data using Cell Ranger and Seurat, bioinformaticsworkbook.org (useful)
- how to get/make/find fastq file from cellranger?
- filtered_feature_bc_matrix vs raw_feature_bc_matrix. "Filtered feature-barcode matrix" contains only detected cellular barcodes. <raw_feature_bc_matrix> contains mainly empty barcodes. That is, the difference is the number of cells.
- 3 files are created under 'filtered_feature_bc_matrix' directory: barcodes.tsv.gz, features.tsv.gz, matrix.mtx.gz
- Using filtered_feature_bc_matrix for scRNAseq samples
- https://hpc.nih.gov/apps/cellranger.html
- Reference genome
module load cellranger/5.0.0 echo $CELLRANGER_REF/refdata-gex-GRCh38-2020-A # /fdb/cellranger/refdata-gex-GRCh38-2020-A ls /fdb/cellranger/refdata-gex-GRCh38-2020-A # fasta genes pickle reference.json star ls /fdb/cellranger/ # refdata-cellranger-1.1.0 refdata-cellranger-GRCh38-1.2.0 refdata-cellranger-vdj-GRCh38-alts-ensembl-2.0.0 # refdata-cellranger-1.2.0 refdata-cellranger-GRCh38-3.0.0 refdata-cellranger-vdj-GRCh38-alts-ensembl-3.1.0 # refdata-cellranger-2.0.0 refdata-cellranger-GRCh38-and-mm10-3.1.0 refdata-cellranger-vdj-GRCh38-alts-ensembl-4.0.0 # refdata-cellranger-2020-A refdata-cellranger-hg19-1.2.0 refdata-cellranger-vdj-GRCh38-alts-ensembl-5.0.0 # refdata-cellranger-2.1.0 refdata-cellranger-hg19-3.0.0 refdata-cellranger-vdj-GRCm38-alts-ensembl-2.2.0 # refdata-cellranger-2.2.0 refdata-cellranger-hg19_and_mm10-1.2.0 refdata-cellranger-vdj-GRCm38-alts-ensembl-3.1.0 # refdata-cellranger-3.0.0 refdata-cellranger-hg19_and_mm10-2.1.0 refdata-cellranger-vdj-GRCm38-alts-ensembl-4.0.0 # refdata-cellranger-3.1.0 refdata-cellranger-hg19-and-mm10-3.0.0 refdata-cellranger-vdj-GRCm38-alts-ensembl-5.0.0 # refdata-cellranger-4.0.0 refdata-cellranger-mm10-1.2.0 refdata-gex-GRCh38-2020-A # refdata-cellranger-5.0.0 refdata-cellranger-mm10-2.1.0 refdata-gex-GRCh38_and_mm10-2020-A # refdata-cellranger-ercc92-1.2.0 refdata-cellranger-mm10-3.0.0 refdata-gex-mm10-2020-A ls /fdb/cellranger/refdata-gex-GRCh38-2020-A/genes # genes.gtf wc -l /fdb/cellranger/refdata-gex-GRCh38-2020-A/genes/genes.gtf # 2765974 /fdb/cellranger/refdata-gex-GRCh38-2020-A/genes/genes.gtf # One column contains "gene_name" head -n 6 /fdb/cellranger/refdata-gex-GRCh38-2020-A/genes/genes.gtf ##description: evidence-based annotation of the human genome (GRCh38), version 32 (Ensembl 98) ##provider: GENCODE ##contact: [email protected] ##format: gtf ##date: 2019-09-05 chr1 HAVANA gene 29554 31109 . + . gene_id "ENSG00000243485"; gene_version "5"; gene_type "lncRNA"; gene_name "MIR1302-2HG"; level 2; hgnc_id "HGNC:52482"; tag "ncRNA_host"; havana_gene "OTTHUMG00000000959.2";
- cellranger mkfastq wraps Illumina's bcl2fastq to correctly demultiplex Chromium-prepared sequencing samples and to convert barcode and read data to FASTQ files.
- cellranger count takes FASTQ files from cellranger mkfastq and performs alignment, filtering, and UMI counting. It uses the Chromium cellular barcodes to generate gene-cell matrices and perform clustering and gene expression analysis.
- cellranger aggr aggregates results from cellranger count. How does cellranger aggr normalize for sequencing depth among multiple libraries?
- cellranger reanalyze takes feature-barcode matrices produced by cellranger count or aggr and re-runs the dimensionality reduction, clustering, and gene expression algorithms.
- The example code will generate an output directory with
$ tree s1/outs ├── analysis │ ├── clustering │ │ ├── graphclust │ │ │ └── clusters.csv │ │ ├── kmeans_10_clusters │ │ │ └── clusters.csv │ │ ├── kmeans_2_clusters │ │ │ └── clusters.csv │ │ ├── kmeans_3_clusters │ │ │ └── clusters.csv │ │ ├── kmeans_4_clusters │ │ │ └── clusters.csv │ │ ├── kmeans_5_clusters │ │ │ └── clusters.csv │ │ ├── kmeans_6_clusters │ │ │ └── clusters.csv │ │ ├── kmeans_7_clusters │ │ │ └── clusters.csv │ │ ├── kmeans_8_clusters │ │ │ └── clusters.csv │ │ └── kmeans_9_clusters │ │ └── clusters.csv │ ├── diffexp │ │ ├── graphclust │ │ │ └── differential_expression.csv │ │ ├── kmeans_10_clusters │ │ │ └── differential_expression.csv │ │ ├── kmeans_2_clusters │ │ │ └── differential_expression.csv │ │ ├── kmeans_3_clusters │ │ │ └── differential_expression.csv │ │ ├── kmeans_4_clusters │ │ │ └── differential_expression.csv │ │ ├── kmeans_5_clusters │ │ │ └── differential_expression.csv │ │ ├── kmeans_6_clusters │ │ │ └── differential_expression.csv │ │ ├── kmeans_7_clusters │ │ │ └── differential_expression.csv │ │ ├── kmeans_8_clusters │ │ │ └── differential_expression.csv │ │ └── kmeans_9_clusters │ │ └── differential_expression.csv │ ├── pca │ │ └── 10_components │ │ ├── components.csv │ │ ├── dispersion.csv │ │ ├── features_selected.csv │ │ ├── projection.csv │ │ └── variance.csv │ ├── tsne │ │ └── 2_components │ │ └── projection.csv │ └── umap │ └── 2_components │ └── projection.csv ├── cloupe.cloupe ├── filtered_feature_bc_matrix │ ├── barcodes.tsv.gz │ ├── features.tsv.gz │ └── matrix.mtx.gz # 36601 x 25 after dim(Read10X(data.dir ="filtered_feature_bc_matrix")) ├── filtered_feature_bc_matrix.h5 ├── metrics_summary.csv ├── molecule_info.h5 ├── possorted_genome_bam.bam ├── possorted_genome_bam.bam.bai ├── raw_feature_bc_matrix │ ├── barcodes.tsv.gz │ ├── features.tsv.gz │ └── matrix.mtx.gz # 36601 x 737280 ├── raw_feature_bc_matrix.h5 └── web_summary.html 31 directories, 41 files
# The 2nd column contains gene symbol $ zcat s1/outs/filtered_feature_bc_matrix/features.tsv.gz | head -5 ENSG00000243485 MIR1302-2HG Gene Expression ENSG00000237613 FAM138A Gene Expression ENSG00000186092 OR4F5 Gene Expression ENSG00000238009 AL627309.1 Gene Expression ENSG00000239945 AL627309.3 Gene Expression # R > x <- Read10X("s1/outs/filtered_feature_bc_matrix") > dim(x) [1] 36601 25 > x[1:5, 1:3] 5 x 3 sparse Matrix of class "dgCMatrix" AACGTTGGTTAAAGTG-1 AGTAGTCAGAGCTATA-1 ATCTGCCCATACTCTT-1 MIR1302-2HG . . . FAM138A . . . OR4F5 . . . AL627309.1 . . . AL627309.3 . . . > length(unique(rownames(x))) [1] 36601
- Reference genome
- Why do we have Barcodes with Counts that are not Called Cells? Ambient RNA, A barcode is called a cell if Total UMI count > M/10 is called a cell, M = Cell with maximum total UMI counts (99th percentile of the top N)
- Reads are considered duplicated, if they map to the same gene and have the same UMI. Instead of counting reads we will count number of unique UMIs per gene.
10x Genomics fragment schematic
10x Genomics fragment schematic
10x Molecule Info. This is referenced by the DropletUtils::read10xMolInfo() function.
plot
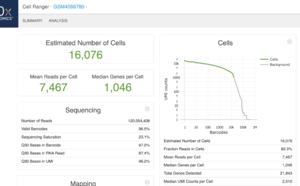
This is an example (GSM4088780) of part of "web_summary.html" output from running cellranger count 2.1.1.
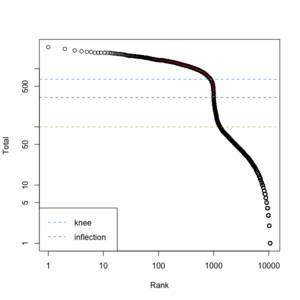
We can create a similar plot using the DropletUtils::barcodeRanks() function. Here is the plot from running the example.
- 11,100 barcoded droplets
- totals =colSums(mat). It has 877 unique values. Range is [0, 2344].
- lower=100. Only cells satisfies the cutoff will be used in fitting the curve and find left.edge, right.edge, knee, inflection.
- run.totals 877 values. Range [0, 2344]
- run.rank 877 values. Range [1, 10900.5]
- out = 11,100 x 3. Columns include rank, total, fitted
- left.edge=68, rigth.edge=653. Associate with the x-axis. Relative to x, y.
- knee=657, inflection=321 (inflection <- 10^(y[right.edge])). Used in the y-axis
- In cellranger count, y-axis = UMI counts, x-axis = Barcodes.
- In cellranger count, green points = cells, gray points = background.
emptyDrops() and DropletQC
- Identify likely cell-containing droplets.
- It returns FDR for each barcoded droplets. Users define a cutoff to subset the matrix to the cell-containing droplets.
- EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data, source code
set.seed(0) my.counts <- DropletUtils:::simCounts() # Identify likely cell-containing droplets. out <- emptyDrops(my.counts) out # DataFrame with 11100 rows and 5 columns # Total LogProb PValue Limited FDR # <integer> <numeric> <numeric> <logical> <numeric> # 1 2 NA NA NA NA # 2 9 NA NA NA NA # ... ... ... ... ... ... # 11099 191 -228.763 9.999e-05 TRUE 0.00013799 # 11100 198 -233.043 9.999e-05 TRUE 0.00013799 is.cell <- out$FDR <= 0.01 sum(is.cell, na.rm=TRUE) # 943
DropletQC – improved identification of empty droplets and damaged cells in single-cell RNA-seq data
estimateAmbience()
This function obtains an estimate of the composition of the ambient pool of RNA based on the barcodes with total UMI counts less than or equal to "lower". The default approach is to assume that all barcodes with total counts less than or equal to "lower" are empty.
It returns a numeric vector of length equal to nrow(m), containing the estimated the transcript proportions in the ambient solution.
set.seed(0) my.counts <- DropletUtils:::simCounts() dim(my.counts) # [1] 100 11100 ambience <- estimateAmbience(my.counts) head(ambience) # GENE1 GENE2 GENE3 GENE4 GENE5 GENE6 # 0.0002213677 0.0003984648 0.0006331183 0.0008146427 0.0010935705 0.0012042561 length(ambience) # [1] 100
removeAmbience()
Estimate and remove the ambient profile from a count matrix, given pre-existing groupings of similar cells. This function is largely intended for plot beautification rather than real analysis.
It returns a numeric matrix-like object of the same dimensions as y, containing the counts after removing the ambient contamination.
ngenes <- 1000 set.seed(1) ambient <- runif(ngenes, 0, 0.1) # length = 1000 cells <- c(runif(100) * 10, integer(900)) # length = 1000 y <- matrix(rpois(ngenes * 100, cells + ambient), nrow=ngenes) # Pretending that all cells are in one group, in this example. removed <- removeAmbience(y, ambient, groups=rep(1, ncol(y))) dim(y) # [1] 1000 100 dim(removed) # [1] 1000 100 summary(as.vector(y)) # Min. 1st Qu. Median Mean 3rd Qu. Max. # 0.0000 0.0000 0.0000 0.5423 0.0000 20.0000 summary(as.vector(removed)) # Min. 1st Qu. Median Mean 3rd Qu. Max. # 0.000 0.000 0.000 0.515 0.000 20.000
Use tools other than cellranger
See Chapter 3 Processing Raw scRNA-seq Data and Chapter 4 Construction of expression matrix.
(non-UMI) Kallisto-Bustools or Alevin, featureCounts, RSEM. Note Kallisto can do both alignment and gene counting.
(UMI-based) UMI-tools & zUMIs, zUMIs: A fast and flexible pipeline to process RNA sequencing data with UMIs
PDX
Seurat*
- http://satijalab.org/seurat/. It has several vignettes.
- https://videocast.nih.gov/summary.asp?Live=21733&bhcp=1
- BingleSeq - A user-friendly R package for Bulk and Single-cell RNA-Seq data analyses.
- Question: Is it worth it to explore outside of Seurat?
- Introduction vignette
Purpose Code Comment Setup the Seurat Object pbmc.data <- Read10X(data.dir = "../data/pbmc3k/filtered_gene_bc_matrices/hg19/")
pbmc <- CreateSeuratObject(counts = pbmc.data, project = "pbmc3k", min.cells = 3, min.features = 200)
pbmc[["RNA"]]@countsNote the @ operator pbmc.data[c("CD3D", "TCL1A", "MS4A1"), 1:30] dense.size <- object.size(as.matrix(pbmc.data)) QC pbmc[["percent.mt"]] <- PercentageFeatureSet(pbmc, pattern = "^MT-")
VlnPlot(pbmc, features = c("nFeature_RNA", "nCount_RNA", "percent.mt"), ncol = 3)
plot1 <- FeatureScatter(pbmc, feature1 = "nCount_RNA", feature2 = "percent.mt")
plot2 <- FeatureScatter(pbmc, feature1 = "nCount_RNA", feature2 = "nFeature_RNA")
plot1 + plot2
pbmc <- subset(pbmc, subset = nFeature_RNA > 200 & nFeature_RNA < 2500 & percent.mt < 5)select cells with number of features in the range of (200,2500) Normalizing the data pbmc <- NormalizeData(pbmc, normalization.method = "LogNormalize", scale.factor = 10000)
pbmc[["RNA"]]@dataNote the @ operator feature selection pbmc <- FindVariableFeatures(pbmc, selection.method = "vst", nfeatures = 2000)
top10 <- head(VariableFeatures(pbmc), 10)
plot1 <- VariableFeaturePlot(pbmc)
plot2 <- LabelPoints(plot = plot1, points = top10, repel = TRUE)
plot1 + plot2Scaling the data all.genes <- rownames(pbmc)
pbmc <- ScaleData(pbmc, features = all.genes)
pbmc[["RNA"]]@scale.dataNote the @ operator Perform linear dimensional reduction pbmc <- RunPCA(pbmc, features = VariableFeatures(object = pbmc))
print(pbmc[["pca"]], dims = 1:5, nfeatures = 5)
VizDimLoadings(pbmc, dims = 1:2, reduction = "pca")
DimPlot(pbmc, reduction = "pca")
DimHeatmap(pbmc, dims = 1, cells = 500, balanced = TRUE)
DimHeatmap(pbmc, dims = 1:15, cells = 500, balanced = TRUE)Determine the ‘dimensionality’ of the dataset pbmc <- JackStraw(pbmc, num.replicate = 100)
pbmc <- ScoreJackStraw(pbmc, dims = 1:20)Determine the ‘dimensionality’ of the dataset pbmc <- JackStraw(pbmc, num.replicate = 100)
pbmc <- ScoreJackStraw(pbmc, dims = 1:20)
JackStrawPlot(pbmc, dims = 1:15)
ElbowPlot(pbmc)Cluster the cells pbmc <- FindNeighbors(pbmc, dims = 1:10)
pbmc <- FindClusters(pbmc, resolution = 0.5)
head(Idents(pbmc), 5)Non-linear transformation: UMAP/tSNE pbmc <- RunUMAP(pbmc, dims = 1:10)
DimPlot(pbmc, reduction = "umap")Finding differentially expressed features cluster1.markers <- FindMarkers(pbmc, ident.1 = 2, min.pct = 0.25)
head(cluster1.markers, n = 5)
cluster5.markers <- FindMarkers(pbmc, ident.1 = 5, ident.2 = c(0, 3), min.pct = 0.25)
head(cluster5.markers, n = 5)
pbmc.markers <- FindAllMarkers(pbmc, only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25)
pbmc.markers %>% group_by(cluster) %>% top_n(n = 2, wt = avg_log2FC)
cluster1.markers <- FindMarkers(pbmc, ident.1 = 0, logfc.threshold = 0.25, test.use = "roc", only.pos = TRUE)
VlnPlot(pbmc, features = c("MS4A1", "CD79A"))
VlnPlot(pbmc, features = c("NKG7", "PF4"), slot = "counts", log = TRUE)
FeaturePlot(pbmc, features = c("MS4A1", "GNLY", "CD3E", "CD14", "FCER1A", "FCGR3A", "LYZ", "PPBP", "CD8A"))
top10 <- pbmc.markers %>% group_by(cluster) %>% top_n(n = 10, wt = avg_log2FC)
DoHeatmap(pbmc, features = top10$gene) + NoLegend()Assigning cell type identity to clusters new.cluster.ids <- c("Naive CD4 T", "CD14+ Mono", "Memory CD4 T", "B", "CD8 T", "FCGR3A+ Mono", "NK", "DC", "Platelet")
names(new.cluster.ids) <- levels(pbmc)
pbmc <- RenameIdents(pbmc, new.cluster.ids)
DimPlot(pbmc, reduction = "umap", label = TRUE, pt.size = 0.5) + NoLegend()
seurat-wrappers
https://github.com/satijalab/seurat-wrappers
Seurat object
- CreateSeuratObject()
- Seurat DimPlot - Highlight specific groups of cells in different colours. seuratObject$meta.data, integrated@[email protected], slotNames(seuratObject), slot(seuratObject, "meta.data").
- Add a new column of meta data. ?AddMetaData
subset genes
<Method 1>: How to subset on genes and preserve idents?
seurat.object # original seurat object
set.seed(1); genes.use <- sample(1:nrow(seurat.object), 1000)
subset.matrix <- seurat.object[['RNA']]@data[genes.use, ]
seurat.object_sub <- CreateSeuratObject(subset.matrix)
seurat.object_sub <- SetIdent(seurat.object_sub,
value=as.factor([email protected]$manual_cluster_tree_addmodulescore))
<Method 2>: subset() function. See Seurat Command List.
<Method 3>: Seurat Weekly NO.2 || 我该如何取子集?(Downsampling)
Seurat methods
- Seurat Methods
- Seurat Command List
- Reference
- Most important
- [email protected]
- pbmc_small[['RNA']]@counts, pbmc_small[['RNA']]@data, pbmc_small[['RNA']]@scale.data
- pbmc[["pca"]], pbmc[["umap"]]
QC
QC part explanation:
> pbmc <- CreateSeuratObject(counts = pbmc.data, project = "pbmc3k", min.cells = 3, min.features = 200)
> pbmc[["percent.mt"]] <- PercentageFeatureSet(pbmc, pattern = "^MT-")
> pbmc2 <- subset(pbmc, subset = nFeature_RNA > 200 & nFeature_RNA < 2500)
> pbmc
An object of class Seurat
13714 features across 2700 samples within 1 assay
Active assay: RNA (13714 features, 0 variable features)
> pbmc2
An object of class Seurat
13714 features across 2695 samples within 1 assay
Active assay: RNA (13714 features, 0 variable features)
> ind <- which(!colnames(pbmc[['RNA']]) %in% colnames(pbmc2[['RNA']]))
> ind
[1] 427 1060 1762 1878 2568
> tmp <- apply(pbmc[['RNA']]@data, 2, function(x) sum(x > 0)) # detected genes per cell
> range(tmp)
[1] 212 3400
> hist(tmp) # kind of normal
> which(tmp > 2500)
AGAGGTCTACAGCT-1 CCAGTCTGCGGAGA-1 GCGAAGGAGAGCTT-1
427 1060 1762
GGCACGTGTGAGAA-1 TTACTCGAACGTTG-1
1878 2568
> tmp2 <- colSums(pbmc[['RNA']]@data) # total counts per cell
> hist(tmp2) # kind of normal
Normalization
- NormalizeData(). log1p(value/colSums[cell-idx] *scale_factor). The scale_factor is 10,000 by default and this gives a good range on the normalized values (at least on the pbmc data). The cpp source code of LogNorm() in preprocessing.R.
R> range(log1p(pbmc[['RNA']]@data[,1]/sum(pbmc[['RNA']]@data[,1]) * 10000)) [1] 0.000000 5.753142 R> range(log1p(pbmc[['RNA']]@data[,1]/sum(pbmc[['RNA']]@data[,1]) * 1000)) [1] 0.000000 3.478712 R> range(log1p(pbmc[['RNA']]@data[,1]/sum(pbmc[['RNA']]@data[,1]) * 100000)) [1] 0.000000 8.052868
And below is a comparison of the raw counts and normalized values:
R> pbmc2 <- NormalizeData(pbmc, normalization.method = "LogNormalize", scale.factor = 10000) R> cbind(pbmc[['RNA']]@data[i, 1], pbmc2[['RNA']]@data[i, 1]) [,1] [,2] MRPL20 1 1.635873 RPL22 1 1.635873 TNFRSF25 2 2.226555 EFHD2 1 1.635873 NECAP2 1 1.635873 AKR7A2 1 1.635873 CAPZB 1 1.635873 RPL11 41 5.138686 RP5-886K2.3 1 1.635873 PITHD1 1 1.635873
Integration datasets
- Using Seurat with multimodal data
- Integrating scRNA-seq and scATAC-seq data
- FindIntegrationAnchors()
- Cell type abundance analysis in scRNA-seq
DE analysis
- 'samples' was used to mean 'cells' according to the vignette Seurat - Guided Clustering Tutorial
- [email protected]
- Idents(seuratObject)
- seuratObject <- SetIdent(seuratObject, value=as.factor())
- seuratObject <- RenameIdents(seuratObject, new.cluster.ids)
- By default, it (FindAllMarkers()) identifies positive and negative markers of a single cluster (specified in ident.1), compared to all other cells.
- FindMarkers()
- Groups
- The vignettes only give examples about the comparisons between clusters.
- For comparison between samples, see
- How to find the difference gene between samples #325.
- Calculating differential expression genes between two sample sequenced using 10x where the vignette Introduction to scRNA-seq integration was recommended.
- Using Seurat to compare mutant vs.wt
- Pseudobulk differential expression analysis via Bioconductor.
- 12 Differential Expression (DE) analysis from the ebook 'Analysis of single cell RNA-seq data' Davis J. McCarthy
- Bias, robustness and scalability in single-cell differential expression analysis Soneson 2018
- Run in parallel for FindAllMarkers() on a large data (33694 features, 114331 + 41956 cells) will result in different errors I use
- parLapply: Error in unserialize(node$con) : error reading from connection. See Going parallel only intermittently works with the same code. The same error message can be reproduced here: How-to go parallel in R – basics + tips. Using makeCluster(no_cores, type="FORK") is an important tool for handling memory ceilings. As they link to the original variable address the fork will not require any time for exporting variables or take up any additional space when using these.
- mclapply: scheduled cores 4, 6, 8 did not deliver results, all values of the jobs will be affected
- foreach: Error in unserialize(socklist[[n]]) : error reading from connection
- future: No error but one cluster returns no genes. Running FindAllMarkers() with 2 clusters does give an error. See how to adjust future.globals.maxSize in R?
Visualization
- DoHeatmap() (source code) and a modified version called DoMultiBarHeatmap() to include multiple color bars for samples/cells.
Azimuth
Azimuth - App for reference-based single-cell analysis
GUI/web applications
Asc-Seurat – Analytical single-cell Seurat-based web application
tidyseurat
Bioconductor packages
| Description | Packages |
|---|---|
| Orchestrating Single-Cell Analysis with Bioconductor (OSCA) simpleSingleCell is an earlier version |
See the list here |
| single cell RNA-Seq workflow (included in OSCA now) | simpleSingleCell |
| visualization tools for single-cell and Bulk RNA Sequencing | dittoSeq |
| An Introduction to Single-Cell RNA-sequencing Analysis in Bioconductor (中文, based on OSCA ebook) | DropletUtils, scran, scater, SingleR, BiocFileCache |
| An Opinionated Computational Workflow for Single-Cell RNA-seq and ATAC-seq | SingleCellExperiment, DropletUtils, scater, iSEE(shiny), scran, limma, edgeR |
| SplatPop: Simulating Population-Scale Single-Cell RNA-sequencing Data | splatter |
| Importing alevin scRNA-seq counts into R/Bioconductor (check 'Get started') | tximeta, SingleCellExperiment, fishpond, scran, Seurat |
GUI
NIDAP
https://nidap.nih.gov/ which uses Code workbook (Code Workbook is an application that allows users to analyze and transform data using an intuitive graphical interface) for the interaction. The R/python analysis code can be accessed just like GEO2R from GEO. The background material is available on here.
Partek
BingleSeq
BingleSeq: a user-friendly R package for bulk and single-cell RNA-Seq data analysis
How many cells are in the human body?
Power analysis
- powsimR: power analysis for bulk and single cell RNA-seq experiments. It is time-consuming and error-prone to install lots of required packages. Better to have a Docker image.
- SCOPIT: sample size calculations for single-cell sequencing experiments
- POWSC - Simulation, power evaluation and sample size recommendation for single-cell RNA-seq, Su 2020.
- SC2P and the paper Two-phase differential expression analysis for single cell RNA-seq Wu 2018.
library(devtools) install_github("haowulab/SC2P", build_vignettes=TRUE) install_github("suke18/POWSC", build_vignettes = T, dependencies = T) library(POWSC) data("es_mef_sce") sce = es_mef_sce[, colData(es_mef_sce)$cellTypes == "fibro"] est_Paras = Est2Phase(sce) sim_size = c(100, 400, 1000) # A numeric vector pow_rslt = runPOWSC(sim_size = sim_size, est_Paras = est_Paras, per_DE=0.05, DE_Method = "MAST", Cell_Type = "PW") # Note, using our previous developed tool SC2P is faster. packageVersion("POWSC") # [1] '0.1.0' help(package="POWSC") plot.POWSC(pow_rslt, Form="II", Cell_Type = "PW") summary.POWSC(pow_rslt, Form="II", Cell_Type = "PW") # (0,10] (10,20] (20,40] (40,80] (80,160] (160,Inf] # 100 0.0171 0.1270 0.2877 0.2740 0.4909 0.6765 # 400 0.3200 0.5373 0.6486 0.7436 0.7959 0.8429 # 1000 0.5534 0.7424 0.8095 0.9067 0.9815 0.9077
Mitochondrion
- https://en.wikipedia.org/wiki/Mitochondrion
- WHAT IS A CELL SIMILAR TOO? In real life cells, the mitochondria is much like a power generator, giving power and energy to a cell, so that it may carry out its other functions.
- Question: Mitochondrial Gene percentage threshold in single cell RNA-Seq. For example, 30% of total mRNA in the heart is mitochondrial due to high energy needs of cardiomyocytes, compared with 5% or less in tissues with low energy demands. For instance, 30% mitochondrial mRNA is representative of a healthy heart muscle cell, but would represent a stressed lymphocyte.
- Seurat::PercentageFeatureSet()
Spike-in
- RNA spike-in. An RNA spike-in is an RNA transcript of known sequence and quantity used to calibrate measurements in RNA hybridization assays, such as DNA microarray experiments, RT-qPCR, and RNA-Seq.
- scran vignette.
- We perform some cursory quality control to remove cells with low total counts or high spike-in percentages.
- An alternative approach is to normalize based on the spike-in counts. The idea is that the same quantity of spike-in RNA was added to each cell prior to library preparation. Size factors are computed to scale the counts such that the total coverage of the spike-in transcripts is equal across cells.
- If we have spike-ins, we can use them to fit the trend (variance-mean relationship) instead.
- Spike-in gene concentrations are known, normalization model existing technical variation by utilizing the difference between these known values and the values observed after processing. "Normalization Methods on Single-Cell RNA-seq Data: An Empirical Survey". Lytal 2020
CPM and glmpca package
Feature selection and dimension reduction for single-cell RNA-Seq based on a multinomial model, glmpca package. All codes are available in github. A talk by Hicks 2021.
scone package: normalization
Performance Assessment and Selection of Normalization Procedures for Single-Cell RNA-Seq.
Quality control
- Quality control of cells and genes (doublets, ambient, empty drops)
- scRNA-seq-workshop. 由于10x 技术会使得外部RNA,也就是不包含微滴的里的细胞被同时测序。在分析之前,我们需要确保得到的barcode数据都相对应活细胞。这里,我们使用`emptyDrops`来筛选“真”细胞。
- Identifying cell-containing droplets/microwells about emptyDrops.
- Decontamination of ambient RNA in single-cell RNA-seq with DecontX Yang 2020
- scRNAseq-analysis-notes by Ming Tang
- Why do we have Barcodes with Counts that are not Called Cells? Ambient RNA
- Avoiding problems with ambient RNA from OSCA
- A guide to quality controlling your scRNA-Seq data
Doublet
- scDblFinder
- scrublet
- scds
- scrublet
Downsampling
- downsampleMatrix() from the scuttle package
- downsampleMatrix: Downsample a count matrix to a desired proportion
- Source code of downsampleMatrix
R> library(scuttle) R> sce <- mockSCE() R> summary(colSums(counts(sce))) Min. 1st Qu. Median Mean 3rd Qu. Max. 344769 366648 374586 373876 382028 396251 R> downsampled2 <- downsampleMatrix(counts(sce), prop = 10000/colSums(counts(sce)), bycol=TRUE) R> colSums(downsampled2[, 1:5]) Cell_001 Cell_002 Cell_003 Cell_004 Cell_005 10000 10000 10000 10000 10000 - downsampleReads() from the DropletUtils package which imports the scuttle package.
- downsampleReads: Downsamples the reads to produce a matrix of UMI counts, given a HDF5 file containing molecule information
- downsampleReads downsamples reads rather than observed counts.
- (from the help) Subsampling the reads with downsampleReads recapitulates the effect of differences in sequencing depth per cell. This provides an alternative to downsampling with the CellRanger aggr function or subsampling with the 10X Genomics R kit.
- From 10x, How does cellranger aggr normalize for sequencing depth among multiple libraries?
- From 10x, How much sequencing saturation should I aim for? If sequencing saturation is at 50%, it means that every 2 new reads will result in 1 new UMI count (unique transcript) detected. In contrast, 90% sequencing saturation means that 10 new reads are necessary to obtain one new UMI count. If the sequencing saturation is high, additional sequencing would not recover much new information for the library.
- From 10x, How is sequencing saturation calculated?
- Sequencing depth for scRNA-seq.
- Source code of downsampleReads
- A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. After downsampling by total reads, we used write10xCounts from DropletUtils and a custom Python script to generate an HDF5 file for input into our analysis pipelines, as described in the sections that follow and in the ‘Code availability’ section.
- R包DropletUtils使用. 比較 downsampling the UMI count matrix 與 downsampling the raw reads. 假如测序饱和度很高的话,最后的total count 并不会下降太多. It also discussed the use of barcode rank plot, empty droplet and demultiplexing hashed libraries.
- 对10X单细胞reads进行随机抽样. 使用 seqtk 对原始fastq文件进行随机抽样.
R> library(DropletUtils) # Mocking up some 10X HDF5-formatted data. R> out <- DropletUtils:::simBasicMolInfo(tempfile()) R> a <- read10xMolInfo(out) R> names(a) [1] "data" "genes" R> dim(a$data) [1] 9508 5 R> str(a$genes) chr [1:20] "ENSG1" "ENSG2" "ENSG3" "ENSG4" "ENSG5" "ENSG6" "ENSG7" "ENSG8" ... R> a$data[1:3, ] DataFrame with 3 rows and 5 columns cell umi gem_group gene reads <character> <integer> <integer> <integer> <integer> 1 GACT 781899 1 9 10 2 CAGC 747670 1 9 9 3 GTAC 614190 1 6 10 R> unique(a$data$gene) [1] 9 6 15 7 12 13 1 20 4 8 3 14 17 19 18 16 2 5 10 11 R> length(unique(a$data$gene)) # 20 genes [1] 20 R> length(unique(a$data$cell)) # 256 cells [1] 256 # Downsampling by the reads. R> b<- downsampleReads(out, barcode.length=4, prop=0.5) R> dim(b) [1] 20 256 R> b[1:3, 1:5] 3 x 5 sparse Matrix of class "dgCMatrix" AAAA-1 AAAC-1 AAAG-1 AAAT-1 AACA-1 ENSG1 . 1 . 1 5 ENSG2 1 1 1 4 . ENSG3 1 1 . 1 4 R> class(b) [1] "dgCMatrix" attr(,"package") [1] "Matrix" R> 4^4 # ATCG for each character of the barcode [1] 256 R> 20*256 # So the combinations of (cell, gene) are not unique [1] 5120 R> length(unique(a$data$umi)) # So UMIs are unique [1] 9508 R> length(unique(a$data$gem_group)) [1] 1Change prop parameter. Notice the result of downsampling reads does not change much.
R> b1 <- downsampleReads(out, barcode.length=4, prop=.9999) R> colSums(b1)[1:5] AAAA-1 AAAC-1 AAAG-1 AAAT-1 AACA-1 41 37 28 36 35 R> b2 <- downsampleReads(out, barcode.length=4, prop=0.2) R> colSums(b2)[1:5] AAAA-1 AAAC-1 AAAG-1 AAAT-1 AACA-1 37 31 23 31 28 R> b5 <- downsampleReads(out, barcode.length=4, prop=0.5) R> colSums(b5)[1:5] AAAA-1 AAAC-1 AAAG-1 AAAT-1 AACA-1 41 37 28 36 35 R> sum(b1) [1] 9508 R> sum(b2) [1] 8245 R> sum(b5) [1] 9448
- 7.5.2 Downsampling and log-transforming from OSCA
- Down_Sample_Matrix()
- zUMIs
- Current best practices in single-cell RNA-seq analysis: a tutorial. While downsampling throws away data, it also increases technical dropout rates which CPM and other global scaling normalization methods do not. Thus, downsampling can deliver a more realistic representation of what cellular expression profiles would look like at similar count depths.
- A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications Haque 2017. Lots of citations.
- Huang 2020 Evaluation of Cell Type Annotation R Packages on Single-cell RNA-seq Data
- downsample features (genes). down_sample_gene(). The idea is sample(gene_list, size = size, replace = FALSE, prob = prob_list) where gene_list is the list of detected genes and size is 5,000/10,000/15,000 and prob_list is the proportion of gene counts (defined by using rowSums()). We randomly downsampled the features from Fluidigm C1 data into 15,000, 10,000 and 5000 input genes, based on the original log count distribution.
- downsample the reads into 25%, 50%, 75% of the original read depth (with two repetitions) on BAM files, and then realigned files following the method provided by the original manuscript.
Normalization
- Introduction to single-cell RNA-seq. If using a 3’ or 5’ droplet-based method, the length of the gene will not affect the analysis because only the 5’ or 3’ end of the transcript is sequenced. However, if using full-length sequencing, the transcript length should be accounted for.
- Bulk normalization methods
- DESeq2: median of ratios (MoR) approach
- edgeR: trimmed mean of M values
- Count models for normalization of single-cell RNA-seq data. See Single Cell Genomics Day 2021
Cell type deconvolution
- List of distinct cell types in the adult human body
- https://twitter.com/stephaniehicks/status/1358776597264797703?s=20
- single-cell signatures from MSigDB
- CDSeqR: fast complete deconvolution for gene expression data from bulk tissues
Batch correction
- Question: scRNA-seq and the batch effects. The biggest problem I found is, since you have no idea what's the real data suppose to be.
- https://www.biostars.org/p/316204/
- Over-correction can happen.
- Single-cell RNA-seq Analysis on NIDAP - Tutorial Part 2 40:00 (nih network is required). Groups differ by a single experimental modification (eg a knockoff gene or cells of similar lineage used to determine differentiation pattern). However batch correction is necessary in cases such as spike-in cells where we expect these cells should be clustered together.
- Mutual nearest neighbors/MNN. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors Haghverdi 2018...
- One assumption is the batch effect is almost orthogonal to the biological subspace. It also assumes the batch effect variation in much smaller than the biological effect variation between different cell types.
- It is preferable to perform DE analyses using the uncorrected expression values with blocking on the batch, as discussed in Section 11.4... We suggest limiting the use of per-gene corrected values to visualization, e.g., when coloring points on a t-SNE plot by per-cell expression.
- Sadly the R code of the paper does not have the file ('MAP2.csv') and the code does not work anymore (e.g. mnnCorrect() does not have $angles component). Check out the mnnpy module which still has an option to return angles.
- SMNN can eliminate the overcorrection between different cell types & allow that batch effects are non-orthogonal to the biological differences. github source code. Note that the package is only available on github and it depends on the python mnnpy module.
library(reticulate) use_python("/home/rstudio/.conda/envs/sc_env/bin/python") py_config() # confirm library("SMNN") data("data_SMNN") dim(data_SMNN$batch1.mat) # [1] 3000 400 data_SMNN$batch1.mat[1:5, 1:5] dim(data_SMNN$batch2.mat) # [1] 3000 500 # Maker genes markers <- c("Col1a1", "Pdgfra", "Ptprc", "Pecam1") # Corresponding cell type labels for each marker gene cluster.info <- c(1, 1, 2, 3) matched_clusters <- unifiedClusterLabelling(data_SMNN$batch1.mat, data_SMNN$batch2.mat, features.use = markers, cluster.labels = cluster.info, min.perc = 0.3) corrected.results <- SMNNcorrect(batches = list(data_SMNN$batch1.mat, data_SMNN$batch2.mat), batch.cluster.labels = matched_clusters, k=20, sigma=1, cos.norm.in=TRUE, cos.norm.out=TRUE, subset.genes=rownames(data_SMNN$batch2.mat)) # NOTE: subset.genes parameter was added to fix a bug when I'm running the function # [1] "Data preparation ..." # Error: C stack usage 555609078676 is too close to the limit > traceback() 3: py_call_impl(callable, dots$args, dots$keywords) 2: mnnpy$utils$transform_input_data(datas = batches.t, cos_norm_in = cos.norm.in, cos_norm_out = cos.norm.out, var_index = as.character(c(0:ncol(batches.t1))), var_subset = subset.index, n_jobs = n.jobs) 1: SMNNcorrect(batches = list(data_SMNN$batch1.mat, data_SMNN$batch2.mat), batch.cluster.labels = matched_clusters, k = 20, sigma = 1, cos.norm.in = TRUE, cos.norm.out = TRUE, subset.genes = rownames(data_SMNN$batch2.mat)[1:100])
- A benchmark of batch-effect correction methods for single-cell RNA sequencing data Tran 2020
- A multi-center cross-platform single-cell RNA sequencing reference dataset 2021 with the source code on github.
- A multicenter study benchmarking single-cell RNA sequencing technologies using reference samples Chen 2020. github and codeocean (most complete). The reference genome and transcriptome were downloaded from the 10x website as ‘refdata-cellranger-GRCh38-1.2.0.tar.gz’, which corresponds to the GRCh38 genome and the Ensembl version 84 transcriptome.
- Companion paper, source code in github. gene count data (gene_counts_all.rdata).
- Talk (video).
- https://broadinstitute.github.io/2019_scWorkshop/correcting-batch-effects.html
- Seurat (CCA): RunMultiCCA() seems not available any more. Check out Integration and Label Transfer in Seurat 3 and Introduction to scRNA-seq integration FindIntegrationAnchors() & IntegrateData().
# Create 2D UMAP DR plot for data before integration # It can be seen there is a need to do batch effect correction ifnb2 <- ifnb ifnb2 <- NormalizeData(ifnb2) ifnb2 <- FindVariableFeatures(ifnb2, selection.method = "vst", nfeatures = 2000) ifnb2 <- ScaleData(ifnb2, verbose = FALSE) ifnb2 <- RunPCA(ifnb2, npcs = 30, verbose = FALSE) ifnb2 <- RunUMAP(ifnb2, reduction = "pca", dims = 1:30) ifnb2 <- FindNeighbors(ifnb2, reduction = "pca", dims = 1:30) ifnb2 <- FindClusters(ifnb2, resolution = 0.5) # Compared to the 2D plot before and after integration DimPlot(ifnb2, reduction = "umap", group.by = "stim") # not good, ideally two groups # should be mixed together. # It seems the blue color is on top of the salmon color. DimPlot(immune.combined, reduction = "umap", group.by = "stim") # Two groups are mixed. # Compare the cluster plots before and after integration DimPlot(ifnb2, reduction = "umap", split.by = "stim") # not good, ideally the cluster # pattern should be similar in both groups DimPlot(immune.combined, reduction = "umap", split.by = "stim") #good, the cluster # patterns are similar in both groupsFindConservedMarkers() and [email protected], ?FindConservedMarkers. ident.1 & ident.2 are used to define cells from the @active.ident component. For example, we can it to find genes that are conserved markers irrespective of stimulation/group condition in a certain cluster. Together with FeaturePlot() we can see the gene expression in 2D space (one plot per feature). The DotPlot() function with the split.by parameter can be useful for viewing conserved cell type markers across conditions
table([email protected]$stim, [email protected]) table(pbmc_small$groups, [email protected])
- liger LIGER (NMF): liger:: optimizeALS(). Note that in order to use the R package, it is necessary to install hdf5 or libhdf5-dev library. Vignettes are available on github source but not on CRAN package.
- Harmony: RunHarmony()
- Seurat (CCA): RunMultiCCA() seems not available any more. Check out Integration and Label Transfer in Seurat 3 and Introduction to scRNA-seq integration FindIntegrationAnchors() & IntegrateData().
Clustering
- minicore: Fast scRNA-seq clustering with various distance measures
- R语言学习笔记之聚类分析
- A systematic performance evaluation of clustering methods for single-cell RNA-seq data Duò 2018
- Quantify performance using the adjusted Rand Index (ARI), comparing the obtained clustering to the true clusters.
- Evaluates the stability of the clustering results from each method, with respect to random starts. Each method was run five times on each data set (for each k), and we quantify the stability by comparing each pair of such runs using the adjusted Rand Index.
- Comparison of characteristic features across count data sets contains a report comparing the main characteristics of the simulated data sets to those of the underlying Kumor data set.
- The paper contains information about data processing (scater & scran packages) and gene filtering (Seurat and M3Drop packages).
# after I upgrade R from 4.0.x to 4.1.x R> library(DuoClustering2018) snapshotDate(): 2021-05-18 Warning message: DEPRECATION: As of ExperimentHub (>1.17.2), default caching location has changed. Problematic cache: /Users/XXX/Library/Caches/ExperimentHub See https://bioconductor.org/packages/devel/bioc/vignettes/ExperimentHub/inst/doc/ExperimentHub.html#default-caching-location-update
NMF
- https://en.wikipedia.org/wiki/Non-negative_matrix_factorization
- NMF rank度量图该怎么看
- What are the pros and cons of different matrix factorization techniques: Low rank, SVD, and NMF? What are some practical examples of each? In NMF you force the coefficients to be positive.
- What is the difference between non-negative matrix factorization and singular value decomposition? SVD factors contains both positive and negative entries while NMF factors are strictly positive.
cNMF
https://github.com/dylkot/cNMF/ (python)
It takes a count matrix (N cells X G genes) as input and produces a (K x G) matrix of gene expression programs (GEPs) and a (N x K) matrix specifying the usage of each program for each cell in the data.
CancerSubtypes
- CancerSubtypes. ExecuteCNMF() which calls the NMF::nmf() function. Looking at the source code, it seems the 'consensus' is not used by nmf() so it does not affect the predicted groups. It only affect the final 'consensus' slot (matrix).
- CancerSubtypes: an R/Bioconductor package for molecular cancer subtype identification, validation and visualization Xu, 2017.
library(CancerSubtypes)
data(GeneExp)
dim(GeneExp) # 1500 x 100
result=ExecuteCNMF(GeneExp,clusterNum=3,nrun=30)
dim(result$distanceMatrix) # consensus/Similarity returned from nmf()
# [1] 100 100
round(unique(result$distanceMatrix), 3)
# [1] 1.000 0.000 0.167 0.067 0.933 0.100 0.433 0.900 0.833 0.567
#[11] 0.233 0.733 0.267 0.133 0.967 0.633 0.500 0.667 0.200 0.533
sil=silhouette_SimilarityMatrix(result$group, result$distanceMatrix)
plot(sil, col = c("red", "blue", "green"))
# Note that the average silhouette width is computed without considering groups.
# mean(c(.96, .98, 1)) # .98
# mean(sil[, 3]) # 0.9714631 match with the plot
# mean(tapply(sil[, 3], sil[, 1], mean)) # 0.9794998
pca <- prcomp(t(GeneExp))
plot(pca$x[,1], pca$x[, 2], col=result$group, pch=19) # black, red, green
seurat::BuildClusterTree
- https://satijalab.org/seurat/reference/buildclustertree and PlotClusterTree()
- ggtree() was used for plotting. See, scRNA-seq analysis workflow by Roman Hillje - Data Visualization & Bioinformatics
Model
- Zero-inflated model, Compound Poisson distribution, Negative binomial distribution/Gamma-Poisson distribution
- Two-phase models for scRNA-Seq
- Comparison and evaluation of statistical error models for scRNA-seq Choudhary & Satija 2021
DE analysis
GSEA
- twosigma package and the paper TWO-SIGMA-G: A New Competitive Gene Set Testing Framework for scRNA-seq Data Accounting for Inter-Gene and Cell-Cell Correlation Burden et ap.
- fgsea by dpcook
- Integrative differential expression and gene set enrichment analysis using summary statistics for scRNA-seq studies Ma 2020. iDEA package.
AddModuleScore
- Seurat包的打分函数AddModuleScore, Another
- 我们感兴趣的基因,抽出来,每一个细胞算一个这些基因表达的平均值, 背景基因的平均值在于找每个基因的所在的bin,在该bin内随机抽取相应的ctrl个基因作为背景,最后所有的目标基因算一个平均值,所有的背景基因算一个平均值,两者相减就是该gene set 的score值。
- Calculate the average expression levels of each program (cluster) on single cell level, subtracted by the aggregated expression of control feature sets. All analyzed features are binned based on averaged expression, and the control features are randomly selected from each bin.
- 10X空间转录组和10X单细胞数据联合分析方法汇总
- Plot the correlation between number of genes and S score
- Figure 4 in Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions.
- Exploring the inner workings of the AddModuleScore function from the Seurat R package.
- AddModuleScore() meaning
- Identifying cell types in my data? The highest scoring cell type (per cell) is assigned as the cell type. However, according to AddModuleScore to score the cells and assign each cell to the gene set #4365 the scores are not directly comparable but need a normalization step.
- https://github.com/satijalab/seurat/search?q=addmodulescore&type=issues
- ?AddModuleScore. The end result is a new vector ('CD_Features1' in this case) which contains scores for all cells.
- By default 24 bins are created and 100 control features selected from the same bin per analyzed feature.
- It seems AddModuleScore can be used for cell type annotation/identification; see this post and this one. If we have a few cell types, we run AddModuleScore on each cell type (a list of genes for each cell type). For each cell, the cell type with the highest score will be used to represent the cell type.
- It seems AddModuleScore is similar to ssGSEA; for example, they can return a score for each sample/cell and each gene set.
library(Seurat)
data("pbmc_small")
cd_features <- list(c('CD79B', 'CD79A', 'CD19', 'CD180', 'CD200', 'CD3D',
'CD2', 'CD3E', 'CD7', 'CD8A', 'CD14', 'CD1C', 'CD68', 'CD9', 'CD247'))
pbmc_small <- AddModuleScore(
object = pbmc_small,
features = cd_features,
ctrl = 5,
name = 'CD_Features'
)
head(pbmc_small, 3)
orig.ident nCount_RNA nFeature_RNA RNA_snn_res.0.8
ATGCCAGAACGACT SeuratProject 70 47 0
CATGGCCTGTGCAT SeuratProject 85 52 0
GAACCTGATGAACC SeuratProject 87 50 1
letter.idents groups RNA_snn_res.1 CD_Features1
ATGCCAGAACGACT A g2 0 0.4420928
CATGGCCTGTGCAT A g1 0 0.4285719
GAACCTGATGAACC B g2 0 0.9217770
Tweedieverse
Cell type annotation
- How many cell types exist in the human body: 200
- Evaluation of Cell Type Annotation R Packages on Single-cell RNA-seq Data Huang. Source code.
- Seurat vignette Mapping and annotating query datasets. FindTransferAnchors() + TransferData() + AddMetaData()
- SingleR (Bioconductor). This method assigns labels to cells based on the reference samples (e.g. Immgen reference dataset for mouse) with the highest Spearman rank correlations, using only the marker genes between pairs of labels to focus on the relevant differences between cell types. See Chapter 12: Cell type annotation of OSCA.
- The vignette uses reference data from the celldex (Reference Index for Cell Types) package.
- SingleR identifies marker genes from the reference and uses them to compute assignment scores (based on the Spearman correlation across markers) for each cell in the test dataset against each label in the reference. The label with the highest score is the assigned to the test cell, possibly with further fine-tuning to resolve closely related labels.
- Use rownames(obj) to make sure the gene names are comparable from the test and reference datasets.
- SingleR() has several arguments to control the choice of marker genes used for annotation; see trainSingleR().
- Old source code in github
- Single-cell mapper (scMappR) – using scRNA-seq to infer the cell-type specificities of differentially expressed genes
- scSorter: assigning cells to known cell types according to marker genes
- SCSA: A Cell Type Annotation Tool for Single-Cell RNA-seq Data Cao 2020
- Automated methods for cell type annotation on scRNA-seq data Pasquini 2021
- CALLR: a semi-supervised cell-type annotation method for single-cell RNA sequencing data Wei 2021
Cell-level metadata
Cell-level metadata are indispensable for documenting single-cell sequencing datasets Puntambekar 2021. Shiny app. Source code.
GSE137710 is a good example.
Cell subtypes
Some cell types
- B cell, https://askabiologist.asu.edu/b-cell. B淋巴球(B lymphocyte), 屬於後天免疫系統的體液免疫,作用為分泌抗體.
- T cell, https://askabiologist.asu.edu/t-cell. T細胞 是淋巴細胞的一種,在免疫反應中扮演著重要的角色
- Schwann cell 神經膜細胞
- Myelocyte /Myeloid cell 骨髓細胞 bone-marrow cell
- Neuroendocrine cell 神經分泌細胞
- Fibroblast 纖維母細胞
- Endothelium 內皮
Cell-cell interactions
Integrating datasets
- MultiAssayExperiment package.
- Iterative single-cell multi-omic integration using online learning Gao 2021. Online integrative non-negative matrix factorization (iNMF).
- Integrated analysis of multimodal single-cell data Hao 2021
- Muon: multimodal omics analysis framework
- A comparison of data integration methods for single-cell RNA sequencing of cancer samples Richards 2021
scanpy python package
- scanpy
- Preprocessing and clustering 3k PBMCs which save the data as the h5ad' (hdf5) format.
- Comparison of Scanpy-based algorithms to remove the batch effect from single-cell RNA-seq data
- mnnpy
Trajectory/pseudotime analysis
simulation
splatSimulatePaths() from splatter
monocle package for pseudotime analysis
- What is a pseudotime? Pseudotime is a measure of how much progress an individual cell has made through a process such as cell differentiation.
- According to a table on here, pseudotime is a continuous variable.
- This is used by the paper Normalization by distributional resampling of high throughput single-cell RNA-sequencing data Brown 2021 to identify genes with significantly variable expression over pseudo-time.
Spatial Transcriptomics Data Analysis
sincell package
SCell
SCell – integrated analysis of single-cell RNA-seq data
GEVM
Detection of high variability in gene expression from single-cell RNA-seq profiling. Two mouse scRNA-seq data sets were obtained from Gene Expression Omnibus (GSE65525 and GSE60361).
NMFEM
Detecting heterogeneity in single-cell RNA-Seq data by non-negative matrix factorization
Scater
Pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R
NDRindex
NDRindex: a method for the quality assessment of single-cell RNA-Seq preprocessing data
DEsingle
DEsingle – detecting three types of differential expression in single-cell RNA-seq data
totalVI
Joint probabilistic modeling of single-cell multi-omic data with totalVI
Copy number by infercnv
- https://github.com/broadinstitute/inferCNV/wiki
- infercnv - Infer Copy Number Variation from Single-Cell RNA-Seq Data
- We need to install jags software before we install the required package rjags. It seems OK to install this old version 4.3.0 of software on macOS Catalina 10.15.7. To install jags, right click the file 'JAGS-4.3.0.mpkg' and choose 'open'. Then follow the instruction of the installer to complete the installation.
- https://www.jianshu.com/c/4c982303e19f?order_by=top
- It seems the goal is to use the CNV data to cluster cells... All cells that clustered together with spiked in controls were labeled "nontumor" whereas the remaining clusters were labels as 'tumor'.
- 使用inferCNV分析单细胞转录组中拷贝数变异. Or follow the help of plot_cnv(), infercnv::run().
- CNS图表复现13—使用inferCNV来区分肿瘤细胞的恶性与否. 因为10X技术出来的单个细胞的reads数量太少,检测到的基因数量太少,但是inferCNV更新后有一个参数是cutoff ,就很清楚的指出来了:cutoff=1 works well for Smart-seq2, and cutoff=0.1 works well for 10x Genomics 说明它应用于10X单细胞转录组数据是没有问题的。
- 是否是免疫细胞很容易区分那是否是肿瘤细胞呢?
- Question: where to download the gene position file? The infercnv_genes_example object has 10338 genes already
Simpler plot
data(infercnv_data_example) # data.frame 8252 x 20, genes x samples
data(infercnv_annots_example) # data.frame 20 x 1, 10 normal and 10 tumor
data(infercnv_genes_example) # data.frame 10338 x 3
infercnv_genes_example[1:3, ]
# V2 V3 V4
# WASH7P chr1 14363 29806
# LINC00115 chr1 761586 762902
# NOC2L chr1 879584 894689
infercnv_object_example <- infercnv::CreateInfercnvObject(raw_counts_matrix=infercnv_data_example,
gene_order_file=infercnv_genes_example,
annotations_file=infercnv_annots_example,
ref_group_names=c("normal"))
infercnv_object_example <- infercnv::run(infercnv_object_example,
cutoff=1,
out_dir=tempfile(),
cluster_by_groups=TRUE,
denoise=TRUE,
HMM=FALSE,
num_threads=2)
system("ls /var/folders/2q/slryb0rx4tj97t66v7l6pwvr_z6g3s/T//RtmpEuGgVy/file182d24b626381")
# 01_incoming_data.infercnv_obj infercnv.21_denoised.observations_dendrogram.txt
# 02_reduced_by_cutoff.infercnv_obj infercnv.21_denoised.png
# 03_normalized_by_depth.infercnv_obj infercnv.21_denoised.references.txt
# 04_logtransformed.infercnv_obj infercnv.heatmap_thresholds.txt
# 08_remove_ref_avg_from_obs_logFC.infercnv_obj infercnv.observation_groupings.txt
# 09_apply_max_centered_expr_threshold.infercnv_obj infercnv.observations.txt
# 10_smoothed_by_chr.infercnv_obj infercnv.observations_dendrogram.txt
# 11_recentered_cells_by_chr.infercnv_obj infercnv.png
# 12_remove_ref_avg_from_obs_adjust.infercnv_obj infercnv.preliminary.heatmap_thresholds.txt
# 14_invert_log_transform.infercnv_obj infercnv.preliminary.observation_groupings.txt
# 15_no_subclustering.infercnv_obj infercnv.preliminary.observations.txt
# 21_denoise.NF_NA.SD_1.5.NL_FALSE.infercnv_obj infercnv.preliminary.observations_dendrogram.txt
# expr.infercnv.21_denoised.dat infercnv.preliminary.png
# expr.infercnv.dat infercnv.preliminary.references.txt
# expr.infercnv.preliminary.dat infercnv.references.txt
# infercnv.21_denoised.heatmap_thresholds.txt preliminary.infercnv_obj
# infercnv.21_denoised.observation_groupings.txt run.final.infercnv_obj
# infercnv.21_denoised.observations.txt
# Optional, what's the purpose of this plot function
# when the last command already generated the heatmap?
plot_cnv(infercnv_object_example,
out_dir=tempfile(),
obs_title="Observations (Cells)",
ref_title="References (Cells)",
cluster_by_groups=TRUE,
x.center=1,
x.range="auto",
hclust_method='ward.D',
color_safe_pal=FALSE,
output_filename="infercnv",
output_format="png",
png_res=300,
dynamic_resize=0)
# ...
list.files("/var/folders/2q/slryb0rx4tj97t66v7l6pwvr_z6g3s/T//RtmpEuGgVy/file182d25df0c4ed")
# [1] "infercnv.heatmap_thresholds.txt" "infercnv.observation_groupings.txt"
# [3] "infercnv.observations_dendrogram.txt" "infercnv.observations.txt"
# [5] "infercnv.png" "infercnv.references.txt"
More complicated plot
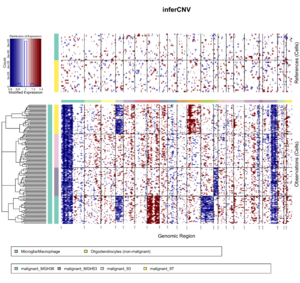
infercnv_obj = CreateInfercnvObject(
raw_counts_matrix=system.file("extdata", "oligodendroglioma_expression_downsampled.counts.matrix.gz", package = "infercnv"),
annotations_file=system.file("extdata", "oligodendroglioma_annotations_downsampled.txt", package = "infercnv"),
delim="\t", gene_order_file=system.file("extdata", "gencode_downsampled.EXAMPLE_ONLY_DONT_REUSE.txt", package = "infercnv"),
ref_group_names=c("Microglia/Macrophage","Oligodendrocytes (non-malignant)"))
infercnv_obj = infercnv::run(infercnv_obj,
cutoff=1, # cutoff=1 works well for Smart-seq2, and cutoff=0.1 works well for 10x Genomics
out_dir=tempfile(),
cluster_by_groups=TRUE,
denoise=TRUE,
HMM=TRUE) # HMM=TRUE will increase lots of computation
# STEP 1: incoming data
# STEP 02: Removing lowly expressed genes
# INFO [2021-08-03 13:38:51] Removing 264 genes from matrix as below mean expr threshold: 1
# INFO [2021-08-03 13:38:51] validating infercnv_obj
# INFO [2021-08-03 13:38:51] There are 5049 genes and 184 cells remaining in the expr matrix.
# STEP 03: normalization by sequencing depth
# ...
# (Error in nls(y ~ .logistic_midpt_slope(x, midpt = x0, slope = k), data = df, : singular gradient
# ), couldn't fit logistic, but no worries, going to use a spline
# (Error in nls(y ~ .logistic_midpt_slope(x, midpt = x0, slope = k), data = df, : singular gradient
# ), couldn't fit logistic, but no worries, going to use a spline
# Error in if (runif(1) <= padj) { : missing value where TRUE/FALSE needed
x <- read.delim(system.file("extdata", "oligodendroglioma_annotations_downsampled.txt", package = "infercnv"), header=F)
dim(x) # [1] 184 2; 184 cells only
unique(x[,2]) # 6 groups. Two of them represent references cells and the other 4 are the obs cells.
# [1] "Microglia/Macrophage" "Oligodendrocytes (non-malignant)"
# [3] "malignant_MGH36" "malignant_MGH53"
# [5] "malignant_97" "malignant_93"
x[1:3, ]
# V1 V2
# 1 MGH54_P2_C12 Microglia/Macrophage
# 2 MGH36_P6_F03 Microglia/Macrophage
# 3 MGH53_P4_H08 Microglia/Macrophage
xx <- read.delim(system.file("extdata", "gencode_downsampled.EXAMPLE_ONLY_DONT_REUSE.txt", package = "infercnv"), header=F)
dim(xx) # [1] 10338 4
xx[1:2, ]
# V1 V2 V3 V4
# 1 WASH7P chr1 14363 29806
# 2 LINC00115 chr1 761586 762902
Cell-cell interactions and communication
Deciphering cell–cell interactions and communication from gene expression Armingol 2021
Chromatin/scATAC-seq data
Signac
Signac is an extension of Seurat for the analysis, interpretation, and exploration of single-cell chromatin datasets.
GPU
NVIDIA Researchers Use AI to Spot Active Areas in Cell DNA
Single Cells and Single Nuclei
- https://en.wikipedia.org/wiki/SnRNA-seq
- Systematic comparison of single-cell and single-nucleus RNA-sequencing methods
- RNA Sequencing from Single Cells and Single Nuclei
- Advantages of single-nucleus over single-cell RNA sequencing