Batch effect
Merging two gene expression studies
Possible batch
STAT115 Chapter 6.5 Batch Effect Removal
Visualization
Chapter 2 Batch effect detection from the ebook Managing Batch Effects in Microbiome Data
ComBat
- Adjusting batch effects in microarray expression data using empirical Bayes methods Johnson 2006. Note the term "ComBat" has not been used.
- Statistics for Genomic Data Science (Coursera) and https://github.com/jtleek/genstats
- Some possible batch variables: operators, runs, machines, library kits, laboratories.
- sva::ComBat() function in sva package from Bioconductor. [math]\displaystyle{ \begin{align} Y_{ijg} = \alpha_g + X \beta_g + \gamma_{ig} + \delta_{ig} \epsilon_{ijg} \end{align} }[/math] where [math]\displaystyle{ X=X_{ij} }[/math] consists of covariates (eg biological) of scientific interests (e.g. Pathway activation levels in Zhang's 2018 simulation example), while [math]\displaystyle{ \gamma_{ig} }[/math] and [math]\displaystyle{ \delta_{ig} }[/math] characterize the additive and multiplicative batch effects of batch i for gene g. The error terms, [math]\displaystyle{ \epsilon_{ijg} }[/math], are assumed to follow a normal distribution with expected value of zero and variance [math]\displaystyle{ \sigma^2_𝑔 }[/math]. The batch corrected data is [math]\displaystyle{ \begin{align} \frac{Y_{ijg} - \hat{\alpha_g} - X \hat{\beta_g} - \hat{\gamma_{ig}}}{\hat{\delta_{ig}}} + \hat{\alpha_g} + X \hat{\beta_g}. \end{align} }[/math]
- Alternative empirical Bayes models for adjusting for batch effects in genomic studies Zhang et al. BMC Bioinformatics 2018. The R package is sva and BatchQC from Bioconductor.
- Reference batch adjustment: [math]\displaystyle{ \begin{align} Y_{ijg} = \alpha_{rg} + X \beta_{rg} + \gamma_{rig} + \delta_{rig} \epsilon_{ijg} \end{align} }[/math] where [math]\displaystyle{ \alpha_{rg} }[/math] is the average gene expression in the chosen reference batch (r). Furthermore, [math]\displaystyle{ \gamma_{rig} }[/math] and [math]\displaystyle{ \delta_{rig} }[/math] represent the additive and multiplicative batch differences between the reference batch and batch i for gene g. The error terms, [math]\displaystyle{ \epsilon_{ijg} }[/math], are assumed to follow a normal distribution with expected value of zero and a reference batch variance [math]\displaystyle{ \sigma^2_{𝑟𝑔} }[/math].
- Mean-only adjustment for batch effects: [math]\displaystyle{ \begin{align} Y_{ijg} = \alpha_{g} + X \beta_{g} + \gamma_{ig} + \epsilon_{ijg} \end{align} }[/math]
- svg vignette example to remove the batch effect
BiocManager::install("sva") library(sva) library(bladderbatch) data(bladderdata) pheno = pData(bladderEset) edata = exprs(bladderEset) batch = pheno$batch table(pheno$cancer) # Biopsy Cancer Normal # 9 40 8 table(batch) # batch # 1 2 3 4 5 # 11 18 4 5 19 modcombat = model.matrix(~1, data=pheno) combat_edata = ComBat(dat=edata, batch=batch, mod=modcombat, prior.plots=FALSE) # This returns an expression matrix, with the same dimensions # as your original dataset (genes x samples). # mod: Model matrix for outcome of interest and other covariates besides batch # By default, it performs parametric empirical Bayesian adjustments. # If you would like to use nonparametric empirical Bayesian adjustments, # use the par.prior=FALSE option (this will take longer). combat_edata = ComBat(dat=edata, batch=batch, ref.batch=1)
- ref.batch for reference-based batch adjustment. mean.only option if there is no need to adjust the variancec. Check out paper Alternative empirical Bayes models for adjusting for batch effects in genomic studies Zhang 2018. Figure 4 shows reference-based ComBat can clearly show the pathway activated samples in Batch 1 samples and show the true data pattern in Batch 2 samples from the simulated study (vs the original ComBat approach failed for both cases). In Figure 5 when we cluster genes using K-means, referenced-based Combat can better identify the role of DE or control genes (compared to the original ComBat method). In addition to the github reposition for the simulation R code, BatchQC::rnaseq_sim() can also do that.
- Merging two gene-expression studies via cross-platform normalization by Shabalin et al, Bioinformatics 2008. This method (called Cross-Platform Normalization/XPN)was used by Ternès Biometrical Journal 2017.
- Batch effect removal methods for microarray gene expression data integration: a survey by Lazar et al, Bioinformatics 2012. The R package is inSilicoMerging which has been removed from Bioconductor 3.4.
- Question: Combine hgu133a&b and hgu133plus2. Adjusting batch effects in microarray expression data using empirical Bayes methods
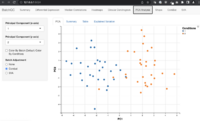
- Figure S1 shows the principal component analysis (PCA) before and after batch effect correction for training and validation datasets from another paper
- Batch effects and GC content of NGS by Michael Love
- 困扰的batch effect
- Some note by Mikhail Dozmorov
CovBat
Mitigating site effects in covariance for machine learning in neuroimaging data Chen 2022. CovBat is a novel harmonization method called Correcting Covariance Batch Effects that removes site effects in mean, variance, and covariance. It was proposed to address the issue of differences in images acquired across multiple sites in multi-site neuroimaging studies. These differences, often referred to as site effects, have been shown to bias comparison between sites, mask biologically meaningful associations, and even introduce spurious associations. The field has focused on harmonizing data by removing site-related effects in the mean and variance of measurements, but these methods may not be sufficient for machine learning. CovBat accounts for the joint distribution of ComBat-adjusted observations.
ComBat-Seq
- sva package vignette
# Example 1 count_matrix <- matrix(rnbinom(400, size=10, prob=0.1), nrow=50, ncol=8) batch <- c(rep(1, 4), rep(2, 4)) adjusted <- ComBat_seq(count_matrix, batch=batch, group=NULL) # Example 2 - one biological variable group <- rep(c(0,1), 4) adjusted_counts <- ComBat_seq(count_matrix, batch=batch, group=group) # Example 3 - multiple biological variables cov1 <- rep(c(0,1), 4) cov2 <- c(0,0,1,1,0,0,1,1) covar_mat <- cbind(cov1, cov2) adjusted_counts <- ComBat_seq(count_matrix, batch=batch, group=NULL, covar_mod=covar_mat)
- ? ComBat_seq It currently (sva version 3.44.0 as of 2022/6) does not have the ref.batch option? See this post Can you add the parameter of ref.batch in ComBat-Seq? #51. One alternative is to normalize the count data of the two batches separately first (for example, using DEseq2) and correct the batch effect then using combat.
- ComBat-seq: batch effect adjustment for RNA-seq count data 2020
- Introduction to Bioconductor SVA and ComBat-Seq in R
- Using ComBat-seq on transcript counts.
- You need to use the RSEM expected counts (values in the expected_counts column are still not integers).
- There is no need to round them to exact integers. You absolutely cannot use TPM or FPKM.
- An example of RSEM output here.
- How to use combat in order to remove batch effects? DESeq() + vst() + limma::removeBatchEffect()
- 数据分析:RNA-seq数据的批次校正方法 It includes complete R code for running ComBat_seq() including ComBat_seq(), limma+removeBatchEffect, PCA, VOOM+SNM, DESeq2.
- Evaluation of critical data processing steps for reliable prediction of gene co-expression from large collections of RNA-seq data Vandenbon 2022
svaseq
Applications
DESeq2
- Batch effects vs biological variables
- Batch-correction on tximport data from salmon. Include the batch effect in the design of DESeq2. No need to use an external tool like ComBat_seq(). Search the keyword batch in the Vignette.
- Batch correction for RNA-seq didn't work with ComBat-seq tool
- How to get TPM / FPKM after batch correction with DESeq2?
- Convert uncorrected counts to FPKM (DESeq2::fpkm()) and then log2 scale and use it for Batch correction with limma::removeBatchEffect.
limma::removeBatchEffect()
- removeBatchEffect() from limma package
- Batch effects : ComBat or removebatcheffects (limma package) ?
- Tutorial:Basic normalization, batch correction and visualization of RNA-seq data
- Extracting fitted expression matrix from Limma
- How to use removeBatchEffect for removing effect of multiple confounding variables? The batch1 and batch2 arguments are really just for convenience. Everything gets combined into the covariates matrix before doing the math.
design <- model.matrix(~TREAT + A + B + C, data=covariates_df) treatment.design <- design[,1:2] batch.design <- design[,-(1:2)] corrected_vst_df <- removeBatchEffect(t(vst_df), design=treatment.design, covariates=batch.design)
ComBat or removebatcheffects (limma package)
- Batch effects : ComBat or removebatcheffects (limma package) ?
- The conclusion that you should get from reading this is that correcting for batch directly with programs like ComBat is best avoided.
- See Methods that remove batch effects while retaining group differences may lead to exaggerated confidence in downstream analyses Nygaard 2016 (215 cites vs 5372 cites from ComBat)
- batch effect : comBat or blocking in limma ?. See the comment by Evan Johnson
- correcting the batch effects in Limma and SVA answered by Gordon Smyth.
ComBat or blocking in limma
batch effect : comBat or blocking in limma ?. The main difference between what Limma does and ComBat is that ComBat adjusts for differences in both the mean and variance differences across the batches, whereas Limma (I believe--Gordon please confirm) assumes that the batch variances are the same and only accounts for mean differences across the batches. So if there are large differences in batch variances, it might still be better to use ComBat. If there are not large variance differences, then Limma should be the best.
Comparisons of svaseq and Combat
Evaluation of Methods in Removing Batch Effects on RNA-seq Data. The results show the SVA method has the best performance, while the ComBat method over-corrects the batch effect.
HarmonizeR, proteomic
MultiBaC- Multiomic Batch effect Correction
BatchQC tool
- https://www.bioconductor.org/packages/release/bioc/html/BatchQC.html
- What is Explained Variance? (Definition & Example)
- The explained variance can be found in the SS (“sum of squares”) column for the Between Groups variation.
- In a regression model, the explained variance is summarized by R-squared, often written R2 = SSR/SST; Coefficient of Determination.
- Some screenshots
AMDBNorm - adjustment mean distribution-based normalization
AMDBNorm: an approach based on distribution adjustment to eliminate batch effects of gene expression data Zhang, Briefings in Bioinformatics 2021
AMDBNorm assumes that each sample in the biologic condition has the same distribution.
It selects a reference batch in advance, then
- aligns other batches with the distribution of the reference batch, and finally
- corrects the gene expression according to their mean values in the reference batch so that the expression of each gene in each batch is at the same level.
Evaluation methods
- Visualization by principal component analysis
- Evaluation of batch effects by BatchQC software
- Principal variance component analysis
- Hierarchical cluster analysis
- Sensitivity analysis of AMDBNorm to reference batch
- Adaptability analysis of batch correction methods to the new samples
TCGA
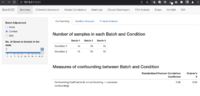
Confounding factors
AC-PCoA: Adjustment for confounding factors using principal coordinate analysis 2022
impute.knn
- ?impute.knn impute.knn(data ,k = 10, rowmax = 0.5, colmax = 0.8, maxp = 1500, rng.seed=362436069)
- rowmax: The maximum percent missing data allowed in any row (default 50%). For any rows with more than rowmax% missing are imputed using the overall mean per sample.
- colmax: The maximum percent missing data allowed in any column (default 80%). If any column has more than colmax% missing data, the program halts and reports an error.
BiocManager::install("impute") library(impute) set.seed(1) x <- matrix(rnorm(10*4), nr=10) x[1,1] <- NA x[2,1:2] <- NA x[3,1:3] <- NA x[1:9, 1] <- NA x2 <- impute.knn(x, k=2) # Error in impute.knn(x, k = 2) : # a column has more than 80 % missing values! set.seed(1) x <- matrix(rnorm(10*4), nr=10) x[1,1] <- NA x[2,1:2] <- NA x[3,1:3] <- NA x2 <- impute.knn(x, k=2) # Warning message: # In knnimp(x, k, maxmiss = rowmax, maxp = maxp) : # 1 rows with more than 50 % entries missing; # mean imputation used for these rows names(x2) [1] "data" "rng.seed" "rng.state" head(x2$data) # [,1] [,2] [,3] [,4] # [1,] 0.1351965 1.51178117 0.91897737 1.35867955 # [2,] -0.5629284 0.27448386 0.78213630 -0.10278773 # [3,] 0.0000000 0.00000000 0.00000000 0.38767161 # [4,] 1.5952808 -2.21469989 -1.98935170 -0.05380504 # [5,] 0.3295078 1.12493092 0.61982575 -1.37705956 set.seed(1) x <- 2^matrix(rnorm(10*4), nr=10) x[1,1] <- NA x[2,1:2] <- NA x[3,1:3] <- NA x2 <- impute.knn(x, k=2) head(x2$data) x3 <- impute.knn(x, k=2) identical(x2$data, x3$data) # trueAnother case that we are able to verify imputed for rows with more than rowmax% missing values. Two rows satisfy the condition. Note the missing value on row 3, column 3 is computed by excluding ALL rows satisfying the condition.
set.seed(1) x <- matrix(rnorm(10*4), nr=10) x[1,1] <- NA x[2,1:2] <- NA x[3,1:3] <- NA x2 <- impute.knn(x, k=2, rowmax=.25) head(x2$data) # [,1] [,2] [,3] [,4] # [1,] 0.1351965 1.51178117 0.91897737 1.35867955 # [2,] 0.3714953 0.33998088 0.78213630 -0.10278773 # [3,] 0.3714953 0.33998088 -0.27417921 0.38767161 # [4,] 1.5952808 -2.21469989 -1.98935170 -0.05380504 mean(x[,1], na.rm = T) # [1] 0.3714953 # same mean(x[,2], na.rm = T) # [1] 0.3399809 # same mean(x[,3], na.rm = T) # [1] -0.1568108 # not the same mean(x[-c(2,3),3], na.rm = T) # [1] -0.2741792 # same