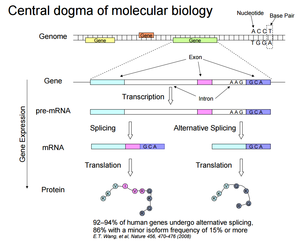
Genome
Visualization
See also Bioconductor > BiocViews > Visualization. Search 'genom' as the keyword.
IGV
nano ~/binary/IGV_2.3.52/igv.sh # Change -Xmx2000m to -Xmx4000m in order to increase the memory to 4GB ~/binary/IGV_2.3.52/igv.sh
Gviz
ChromHeatMap
Heat map plotting by genome coordinate.
ggbio
NOISeq package
Exploratory analysis (Sequencing depth, GC content bias, RNA composition) and differential expression for RNA-seq data.
rtracklayer
R interface to genome browsers and their annotation tracks
- Retrieve annotation from GTF file and parse the file to a GRanges instance. See the 'Counting reads with summarizeOverlaps' vignette from GenomicAlignments package.
ssviz
A small RNA-seq visualizer and analysis toolkit. It includes a function to draw bar plot of counts per million in tag length with two datasets (control and treatment).
Sushi
See fig on p22 of Sushi vignette where genes with different strands are shown with different directions when plotGenes() was used. plotGenes() can be used to plot gene structures that are stored in bed format.
cBioPortal
The cBioPortal for Cancer Genomics provides visualization, analysis and download of large-scale cancer genomics data sets.
https://github.com/cBioPortal/cbioportal
Copy Number
Copy number work flow using Bioconductor
Detect copy number variation (CNV) from the whole exome sequencing
- http://www.ncbi.nlm.nih.gov/pubmed/24172663
- http://www.biomedcentral.com/1471-2164/15/661
- http://www.blog.biodiscovery.com/2014/06/16/comparison-of-cnv-detection-from-whole-exome-sequencing-wes-as-compared-with-snp-microarray/
- exomeCopy package
- exomeCNV package and more info including slides by Fah Sathirapongsasuti.
Whole exome sequencing != whole genome sequencing
NGS
Introduction to Sequence Data Analysis
- Review of RNA-Seq Data Analysis Tools
- RNA-Seq workshop from NYU
- Modern RNA-seq differential expression analyses: transcript-level or gene-level
- http://www.pasteur.fr/~tekaia/BCGA2014/TALKS/Pabinger_Tools4VariantAnalysisOfNGS_EMBO2014.pdf (QC, duplicates, SAM, BAM, picard, variant calling, somatic variant, structural variants, CNV, variant filter, variant annotation, tools for vcf, visualization, pipeline). The source code is available on github.
- https://en.wiki2.org/wiki/List_of_RNA-Seq_bioinformatics_tools
- http://www.rnaseq.wiki/ or https://github.com/griffithlab/rnaseq_tutorial/wiki from The Griffith Lab. It has a paper too at PLOS.
- http://bioinformatics.ca/workshops/2015/high-throughput-biology-sequence-networks-2015#material
- http://www.cbcb.umd.edu/~hcorrada/CMSC858B/
- http://watson.nci.nih.gov/~sdavis/tutorials/biowulf-2011/
- http://cbsu.tc.cornell.edu/ngw2010/Day3_lecture1.pdf
- http://static.msi.umn.edu/tutorial/lifescience/RNA-Seq-Module-1.pdf
- http://www.rna-seqblog.com/introduction-to-rna-seq-analysis/ (focus on statistical part)
- BroadE: GATK/Mapping and processing RNAseq from youtube.
- Low-count transcripts in RNA-Seq analysis pipeline
NIH only
- Some training material from NIH:
- Biowulf seminar by Steven Fellini & Susan Chacko.
- Bash Scripting and Linux commands
Why Do You Need To Use Cdna For Rna-Seq?
https://www.biostars.org/p/54969/
Workshops
- UC Davis Bioinformatics Core.
Automation
- Implementation of an Open Source Software solution for Laboratory Information Management and automated RNA-seq data analysis
- TRAPLINE – a standardized and automated pipeline for RNA sequencing data analysis, evaluation and annotation
Quality control
For base quality score, the quality value, Q(sanger) = - 10log10(prob) where prob = probability that the call at base b is correct. Sanger (Phred quality scores) is between 0 and 93. In practice the maximum quality score is ~40. Quality values below 20 are typically considered low.
FastQC
Trim Galore!
QoRTs
A comprehensive toolset for quality control and data processing of RNA-Seq experiments.
Alignment and Indexing
- Systematic evaluation of spliced alignment programs for RNA-seq data
- (Video) http://bioinformatics.ca//files/public/RNA-seq_2014_Montreal_Module2.mp4, http://www.rna-seqblog.com/rna-seq-alignment-and-visualization/
- (Video) https://youtu.be/n77EAk8C1es RNA-SEQ: Mapping to a Reference Genome
Bowtie2
Extremely fast, general purpose short read aligner.
bowtie needs to have an index of the genome in order to perform its alignment functionality. For example, to build a bowtie index against UCSC hg19 (See Getting started with Bowtie 2 -> Indexing a reference genome)
bowtie2-build /data/ngs/public/sequences/hg19/genome.fa hg19
The reference genome index <genome.fa> can be generated by following Sean Davis' instruction. Note that genome sequence indexes (including Bowtie indexes) as well as GTF transcript annotation files for many commonly used reference genomes can be directly downloaded from http://tophat.cbcb.umd.edu/igenomes.shtml. Even the index file can be directly downloaded without going through Bowtie program, bowtie program is still needed by Tophat program where Tophat's job is to align the RNA-seq data to reference genome.
PS: these genome sequence indexes files are quite big; for example 21 GB for hg19.
To run alignment,
bowtie2 -p 4 -x /data/ngs/public/sequences/hg19 XXX.fastq -S XXX.bt2.sam
At the end of alignment, it will show how many (and percent) of reads are aligned 0 times, exactly 1 time, and >1 times.
We can transform them into a bam format
samtools view -dT XXX.fa XXX.bt2.sam > XXX.bt2.bam
and view the bam file
samtools view XXX.bt2.bam
BWA
Used by whole-exome sequencing. For example, http://bib.oxfordjournals.org/content/15/2/256.full and http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3865592/. Whole Exome Analysis.
Creating index file
bwa index HPV_all.fasta
Mapping
bwa mem # display the help bwa mem -t 4 XXX.fa XXX.fastq > XXX.sam more XXX.sam samtools view -dT XXX.fa XXX.sam > XXX.bam # transform to bam format samtools view XXX.bam | more </syntaxhighlight>
And this is a tutorial to use bwa and freebayes to do variant calling by Freebayes's author.
This tutorial is using a whole genome data SRR030257 from SRP001369
Best pipeline for human whole exome sequencing
Tophat
Aligns RNA-Seq reads to the genome using Bowtie/Discovers splice sites. It does so by splitting longer reads into small sections and aligning those to the genome. It then looks for potential splice sites between pairs of sections to construct a final alignment.
Linux part.
$ type -a tophat # Find out which command the shell executes: tophat is /home/mli/binary/tophat $ ls -l ~/binary
Quick test of Tophat program
$ wget http://tophat.cbcb.umd.edu/downloads/test_data.tar.gz $ tar xzvf test_data.tar.gz $ cd ~/tophat_test_data/test_data $ PATH=$PATH:/home/mli/bowtie-0.12.8 $ export PATH $ ls reads_1.fq test_ref.1.ebwt test_ref.3.bt2 test_ref.rev.1.bt2 test_ref.rev.2.ebwt reads_2.fq test_ref.2.bt2 test_ref.4.bt2 test_ref.rev.1.ebwt test_ref.1.bt2 test_ref.2.ebwt test_ref.fa test_ref.rev.2.bt2 $ tophat -r 20 test_ref reads_1.fq reads_2.fq $ # This will generate a new folder <tophat_out> $ ls tophat_out accepted_hits.bam deletions.bed insertions.bed junctions.bed logs prep_reads.info unmapped.bam
TopHat accepts FASTQ and FASTA files of sequencing reads as input. Alignments are reported in BAM files. BAM is the compressed, binary version of SAM43, a flexible and general purpose read alignment format. SAM and BAM files are produced by most next-generation sequence alignment tools as output, and many downstream analysis tools accept SAM and BAM as input. There are also numerous utilities for viewing and manipulating SAM and BAM files. Perhaps most popular among these are the SAM tools (http://samtools.sourceforge.net/) and the Picard tools (http://picard.sourceforge.net/).
Note that if the data is DNA-Seq, we can merely use Bowtie2 or BWA tools since we don't have to worry about splicing.
An example of using Tophat2 (paired end in this case, 5 threads) is
tophat2 --no-coverage-search -p 5 \
-o "Sample1" \
-G ~/iGenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf \
--transcriptome-index=transcriptome_data/known \
~/iGenomes/Homo_sapiens/UCSC/hg19/Sequence/Bowtie2Index/genome \
myfastq_R1.fastq.gz myfastq_R2.fastq.gz
To find out the alignment rate for ALL bam files (eg we have ctrl1, ctrl2, test1, test2 directories and each of them have align_summary.txt file),
grep "overall read mapping rate" */align_summary.txt
Novel transcripts
STAR (Spliced Transcripts Alignment to a Reference)
Optimizing RNA-Seq Mapping with STAR by Alexander Dobin and Thomas R. Gingeras
Its manual is on github. The 2015 paper includes scripts to run STAR.
Note that the readme file says HARDWARE/SOFTWARE REQUIREMENTS:
- x86-64 compatible processors
- 64 bit Linux or Mac OS X
- 30GB of RAM for human genome
See the blog on gettinggeneticsdone.com for a comparison of speed and memory requirement.
In short, the notable increase in speed comes at the price of a larger memory requirement.
To build STAR on Ubuntu (14.04)
wget https://github.com/alexdobin/STAR/archive/STAR_2.4.2a.tar.gz sudo tar -xzf STAR_2.4.2a.tar.gz -C /opt/RNA-Seq/bin cd /opt/RNA-Seq/bin/STAR-STAR_2.4.2a cd source sudo make STAR
Create index folder
STAR --runMode genomeGenerate --runThreadN 11 \
--genomeDir STARindex \
--genomeFastaFiles genome.fa \
--sjdbGTFfile genes.gtf \
--sjdbOverhang 100
where 100 = read length (one side) -1 = 101 - 100.
STARindex folder
brb@T3600 ~/SRP050992 $ ls ~/SRP012607/STARindex/ chrLength.txt chrStart.txt geneInfo.tab SA sjdbList.fromGTF.out.tab chrNameLength.txt exonGeTrInfo.tab Genome SAindex sjdbList.out.tab chrName.txt exonInfo.tab genomeParameters.txt sjdbInfo.txt transcriptInfo.tab brb@T3600 ~/SRP050992 $ cat ~/SRP012607/STARindex/genomeParameters.txt ### STAR --runMode genomeGenerate --runThreadN 11 --genomeDir STARindex --genomeFastaFiles /home/brb/igenomes/Homo_sapiens/UCSC/hg38/Sequence/WholeGenomeFasta/genome.fa --sjdbGTFfile /home/brb/igenomes/Homo_sapiens/UCSC/hg38/Annotation/Genes/genes.gtf --sjdbOverhang 100 versionGenome 20201 genomeFastaFiles /home/brb/igenomes/Homo_sapiens/UCSC/hg38/Sequence/WholeGenomeFasta/genome.fa genomeSAindexNbases 14 genomeChrBinNbits 18 genomeSAsparseD 1 sjdbOverhang 100 sjdbFileChrStartEnd - sjdbGTFfile /home/brb/igenomes/Homo_sapiens/UCSC/hg38/Annotation/Genes/genes.gtf sjdbGTFchrPrefix - sjdbGTFfeatureExon exon sjdbGTFtagExonParentTranscript transcript_id sjdbGTFtagExonParentGene gene_id sjdbInsertSave Basic
Splice junction
- Manual. Search 'sjdbOverhang' (length of the donor/acceptor sequence on each side of the junctions). The --sjdbOverhang is used only at the genome generation step, and tells STAR how many bases to concatenate from donor and acceptor sides of the junctions. If you have 100b reads, the ideal value of --sjdbOverhang is 99, which allows the 100b read to map 99b on one side, 1b on the other side. One can think of --sjdbOverhang as the maximum possible overhang for your reads. See A post.
- For the <SJ.out.tab> format see section 4.4 of STAR manual. The first few lines of the file looks like
$ head SJ.out.tab
chrM 711 764 1 1 1 2 1 44
chrM 717 1968 1 1 1 1 0 43
chrM 759 1189 0 0 1 11 3 30
chrM 795 830 2 2 1 335 344 30
chrM 822 874 1 1 1 1 1 17
chrM 831 917 2 2 1 733 81 38
chrM 831 3135 2 2 1 1 0 30
chrM 846 1126 0 0 1 4 0 50
chrM 876 922 1 1 1 26 9 31
chrM 962 1052 2 2 1 3 0 30
^ ^ ^ ^ ^ ^ ^ ^ ^
| | | | | | | | |
chrom start end strand intron annotated | # of |
0=undef motif # of multi mapped|
1=+ uniquely mapped max spliced alignment overhang
2=- reads
- Tophat. Search overhang, donor/acceptor keywords. Check out <junctions.bed>. Tophat suggest users use this second option (--coverage-search) for short reads (< 45bp) and with a small number of reads (<= 10 million).
Log.final.out
An example of the Log.final.out file
$ cat Log.final.out
Started job on | Jul 21 13:12:11
Started mapping on | Jul 21 13:35:06
Finished on | Jul 21 13:57:42
Mapping speed, Million of reads per hour | 248.01
Number of input reads | 93418927
Average input read length | 202
UNIQUE READS:
Uniquely mapped reads number | 61537064
Uniquely mapped reads % | 65.87%
Average mapped length | 192.62
Number of splices: Total | 15658546
Number of splices: Annotated (sjdb) | 15590438
Number of splices: GT/AG | 15400163
Number of splices: GC/AG | 178602
Number of splices: AT/AC | 14658
Number of splices: Non-canonical | 65123
Mismatch rate per base, % | 0.86%
Deletion rate per base | 0.02%
Deletion average length | 1.61
Insertion rate per base | 0.03%
Insertion average length | 1.40
MULTI-MAPPING READS:
Number of reads mapped to multiple loci | 4962849
% of reads mapped to multiple loci | 5.31%
Number of reads mapped to too many loci | 62555
% of reads mapped to too many loci | 0.07%
UNMAPPED READS:
% of reads unmapped: too many mismatches | 0.00%
% of reads unmapped: too short | 28.70%
% of reads unmapped: other | 0.05%
CHIMERIC READS:
Number of chimeric reads | 0
% of chimeric reads | 0.00%
Note that 65.87 (uniquely mapped) + 5.31 (mapped to multiple loci) + 0.07 (mapped too many loci) + 28.70 (unmapped too short) + 0.05 (unmapped other) = 100
By default, STAR only outputs reads that map to <=10 loci, others are considered "mapped to too many loci". You can increase this threshold by increasing --outFilterMultimapNmax. See here.
RNASequel
RNASequel: accurate and repeat tolerant realignment of RNA-seq reads
Lessons
- Tophat and STAR needs index files. So if we want to run the alignment using multiple nodes, we want to first run the alignment on a single sample first. Then we can run alignment on others samples using multiple nodes.
- The index files of Tophat only depends on the reference genome. However the index files of STAR depends on both the reference genome and the read length of the data.
- BWA does not require index files. So we can directly run alignment on all samples using multiple nodes.
SAMtools
- https://en.wikipedia.org/wiki/SAMtools
- samtools include samtools, bcftools, bgzip, tabix, wgsim and htslib. samtools and bcftools are based on htslib.
- SAMtools: Primer / Tutorial by Ethan Cerami. It covers installing samtools, bcftools, generating simulated reads, align reads, convert sam to bam, sorting & indexing, identifying genomic variants, understand vcf format, visualize reads on a command line.
$ /opt/RNA-Seq/bin/samtools-0.1.19/samtools
Program: samtools (Tools for alignments in the SAM format)
Version: 0.1.19-44428cd
Usage: samtools <command> [options]
Command: view SAM<->BAM conversion
sort sort alignment file
mpileup multi-way pileup
depth compute the depth
faidx index/extract FASTA
tview text alignment viewer
index index alignment
idxstats BAM index stats (r595 or later)
fixmate fix mate information
flagstat simple stats
calmd recalculate MD/NM tags and '=' bases
merge merge sorted alignments
rmdup remove PCR duplicates
reheader replace BAM header
cat concatenate BAMs
bedcov read depth per BED region
targetcut cut fosmid regions (for fosmid pool only)
phase phase heterozygotes
bamshuf shuffle and group alignments by name
To compile the new version (v1.x) of samtools,
git clone https://github.com/samtools/samtools.git git clone https://github.com/samtools/htslib.git cd samtools make
samtools flagstat: Know how many alignment a bam file contains
brb@brb-T3500:~/GSE11209$ /opt/RNA-Seq/bin/samtools-0.1.19/samtools flagstat dT_bio_s.bam 1393561 + 0 in total (QC-passed reads + QC-failed reads) 0 + 0 duplicates 1393561 + 0 mapped (100.00%:-nan%) 0 + 0 paired in sequencing 0 + 0 read1 0 + 0 read2 0 + 0 properly paired (-nan%:-nan%) 0 + 0 with itself and mate mapped 0 + 0 singletons (-nan%:-nan%) 0 + 0 with mate mapped to a different chr 0 + 0 with mate mapped to a different chr (mapQ>=5)
We can see how many reads have multiple alignments by comparing the number of reads and the number of alignments output from samtools flagstat.
brb@brb-T3500:~/GSE11209$ wc -l SRR002062.fastq 13778616 SRR002062.fastq
samtools view
See also Anders2013-sam/bam and http://genome.sph.umich.edu/wiki/SAM
View bam files:
samtools view XXX.bam # No header samtools view -h XXX.bam # include header in SAM output samtools view -H xxx.bam # See the header only. The header may contains the reference genome fasta information.
Note that according to SAM pdf, the header is optional.
Sam and bam files conversion:
/opt/RNA-Seq/bin/samtools-0.1.19/samtools view --help # sam -> bam (need reference genome file) samtools view -bT XXX.fa XXX.bt2.sam > XXX.bt2.bam # bam -> sam samtools view -h -o out.sam in.bam
Subset:
# assuming bam file is sorted & indexed samtools view XXX.bam "chr22:24000000-25000000" | more
less bam files
http://martinghunt.github.io/2016/01/25/less-foo.bam.html
Picard
A set of tools (in Java) for working with next generation sequencing data in the BAM (http://samtools.sourceforge.net) format.
https://github.com/broadinstitute/picard
Note that As of version 2.0.1 (Nov. 2015) Picard requires Java 1.8 (jdk8u66). The last version to support Java 1.7 was release 1.141. Use the following to check your Java version
brb@T3600 ~/github/picard $ java -version java version "1.7.0_101" OpenJDK Runtime Environment (IcedTea 2.6.6) (7u101-2.6.6-0ubuntu0.14.04.1) OpenJDK 64-Bit Server VM (build 24.95-b01, mixed mode)
See my Linux -> JRE and JDK page.
Some people reported errors or complain about memory usage. See this post from seqanswers.com. And HTSeq python program is another option.
sudo apt-get install ant
git clone https://github.com/broadinstitute/picard.git
cd picard
git clone https://github.com/samtools/htsjdk.git
ant -lib lib/ant package-commands # We will see 'BUILD SUCCESSFUL'
# It will create <dist/picard.jar>
SamToFastq
java jvm-args -jar picard.jar PicardCommandName OPTION1=value1 OPTION2=value2... # For example cd ~/github/freebayes/test/tiny java -Xmx2g -jar ~/github/picard/dist/picard.jar SamToFastq \ INPUT=NA12878.chr22.tiny.bam \ FASTQ=tmp_1.fq \ SECOND_END_FASTQ=tmp_2.fq \ INCLUDE_NON_PF_READS=True \ VALIDATION_STRINGENCY=SILENT wc -l tmp_1.fq # 6532 tmp_1.fq wc -l tmp_2.fq # 6532 tmp_2.fq
BEDtools
The bed format is similar to gtf format but with more compact representation. We can use the apt-get method to install it on Linux environment.
bedtools intersect bedtools bamtobed bedtools bedtobam bedtools getfasta bedtools bamtofastq [OPTIONS] -i <BAM> -fq <FASTQ>
For example,
bedtools intersect -wo -a RefSeq.gtf -b XXX.bed | wc -l # in this case every line is one exon bedtools intersect -wo -a RefSeq.gtf -b XXX.bed | cut -f9 | cut -d ' ' -f2 | more bedtools intersect -wo -a RefSeq.gtf -b XXX.bed | cut -f9 | cut -d ' ' -f2 | sort -u | wc -l bedtools intersect -wo -a RefSeq.bed -b XXX.bed | more # one gene is one line with multiple intervals
One good use of bedtools is to find the intersection of bam and bed files (to double check the insertion/deletion/junction reads in bed files are in accepted_hits.bam). See this page
$ bedtools --version bedtools v2.17.0 $ bedtools intersect -abam accepted_hits.bam -b junctions.bed | samtools view - | head -n 3
We can use bedtool to reverse bam files to fastq files. See https://www.biostars.org/p/152956/
Installation
git clone https://github.com/arq5x/bedtools2.git cd bedtools2 make
bamtofastq
cd ~/github/freebayes/test/tiny # Working directory
~/github/samtools/samtools sort -n NA12878.chr22.tiny.bam NA12878.chr22.tiny.qsort
~/github/bedtools2/bin/bamToFastq -i NA12878.chr22.tiny.qsort.bam \
-fq aln.end1.fq \
-fq2 aln.end2.fq 2> bamToFastq.warning.out
## 60 warnings; see <bamToFastq.warning.out>
*****WARNING: Query ST-E00118:53:H02GVALXX:1:1102:1487:23724 is marked as paired, but its mate does not occur next to it in your BAM file. Skipping.
wc -l tmp.out # 60
wc -l aln.end1.fq # 6548
wc -l aln.end2.fq # 6548
# recall tmp_1.fq is 6532
Cufflinks package
Transcriptome assembly and differential expression analysis for RNA-Seq.
Both Cufflinks and Cuffdiff accept SAM and BAM files as input. It is not uncommon for a single lane of Illumina HiSeq sequencing to produce FASTQ and BAM files with a combined size of 20 GB or larger. Laboratories planning to perform more than a small number of RNA-seq experiments should consider investing in robust storage infrastructure, either by purchasing their own hardware or through cloud storage services.
Tuxedo protocol
- Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks by Trapnell, et al 2012.
- https://speakerdeck.com/stephenturner/rna-seq-qc-and-data-analysis-using-the-tuxedo-suite
- bowtie2 - fast alignment
- tophat2 - splice alignment (rna-seq reads, rna are spliced, introns are removed, some reads may span over 2 exons)
- cufflinks - transcript assembly & quantitation
- cuffdiff2 - differential expression
Cufflinks - assemble reads into transcript
Installation
# about 47MB on ver 2.2.1 for Linux binary version wget http://cole-trapnell-lab.github.io/cufflinks/assets/downloads/cufflinks-2.2.1.Linux_x86_64.tar.gz sudo tar xzvf ~/Downloads/cufflinks-2.2.1.Linux_x86_64.tar.gz -C /opt/RNA-Seq/bin/ export PATH=$PATH:/opt/RNA-Seq/bin/cufflinks-2.2.1.Linux_x86_64/ # test cufflinks -h
Cufflinks uses this map (done from Tophat) against the genome to assemble the reads into transcripts.
# Quantifying Known Transcripts using Cufflinks cufflinks -o OutputDirectory/ -G refseq.gtf mappedReads.bam # De novo Transcript Discovery using Cufflinks cufflinks -o OutputDirectory/ mappedReads.bam
The output files are genes.fpkm_tracking, isoforms.fpkm_tracking, skipped.gtf and transcripts.gtf.
It can be used to calculate FPKM.
- https://www.biostars.org/p/11378/
- http://www.partek.com/Tutorials/microarray/User_Guides/UnderstandingReads.pdf
Cuffcompare - compares transcript assemblies to annotation
Cuffmerge - merges two or more transcript assemblies
First create a text file <assemblies.txt>
./dT_bio/transcripts.gtf ./dT_ori/transcripts.gtf ./dT_tech/transcripts.gtf ./RH_bio/transcripts.gtf ./RH_ori/transcripts.gtf ./RH_tech/transcripts.gtf
Then run
cd GSE11209 cuffmerge -g genes.gtf -s genome.fa assemblies.txt
Cuffdiff
Finds differentially expressed genes and transcripts/Detect differential splicing and promoter use.
Cuffdiff takes the aligned reads from two or more conditions and reports genes and transcripts that are differentially expressed using a rigorous statistical analysis.
Follow the tutorial, we can quickly test the cuffdiff program.
$ wget http://cufflinks.cbcb.umd.edu/downloads/test_data.sam $ cufflinks ./test_data.sam $ ls -l total 56 -rw-rw-r-- 1 mli mli 221 2013-03-05 15:51 genes.fpkm_tracking -rw-rw-r-- 1 mli mli 231 2013-03-05 15:51 isoforms.fpkm_tracking -rw-rw-r-- 1 mli mli 0 2013-03-05 15:51 skipped.gtf -rw-rw-r-- 1 mli mli 41526 2009-09-26 19:15 test_data.sam -rw-rw-r-- 1 mli mli 887 2013-03-05 15:51 transcripts.gtf
In real data,
cd GSE11209
cuffdiff -p 5 -o cuffdiff_out -b genome.fa -L dT,RH \
-u merged_asm/merged.gtf \
./dT_bio/accepted_hits.bam,./dT_ori/accepted_hits.bam,./dT_tech/accepted_hits.bam \
./RH_bio/accepted_hits.bam,./RH_ori/accepted_hits.bam,./RH_tech/accepted_hits.bam
N.B.: the FPKM value (<genes.fpkm_tracking>) is generated per condition not per sample. If we want FPKM per sample, we want to specify one condition for each sample.
Pipeline
CummeRbund
Plots abundance and differential expression results from Cuffdiff. CummeRbund also handles the details of parsing Cufflinks output file formats to connect Cufflinks and the R statistical computing environment. CummeRbund transforms Cufflinks output files into R objects suitable for analysis with a wide variety of other packages available within the R environment and can also now be accessed through the Bioconductor website
The tool appears on the paper Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks by Trapnell, et al 2012.
Finding differentially expressed genes
Big pictures
BWA/Bowtie samtools fa ---------> sam ------> sam/bam (sorted indexed, short reads), vcf or tophat Rsamtools GenomeFeatures edgeR (normalization) ---------> --------------> table of counts --------->
Readings
- Introduction to RNA Sequencing and Analysis Kukurba KR, Montgomery SB. (2015) RNA Sequencing and Analysis. Cold Spring Harb Protoc
- RNA-Seq: a revolutionary tool for transcriptomics
- Alignment and visualization from bioinformatics.ca.
- The RNA-seqlopedia from the Cresko Lab of the University of Oregon.
- RNA-Seq Analysis by Simon Andrews.
Technical replicate vs biological replicate
We sequence more DNA from the same DNA "library", which is called a "technical replicate". If we perform a new experiment with a new organism/tissue/population of cells, which is called a "biological replicate".
Youtube videos
- Analyze public dataset by using Galaxy and IGV David Coil from UC Davis Genoem Center
Download the raw fastq data GSE19602 from GEO and uncompress fastq.bz2 to fastq (~700MB) file. NOTE: the data downloaded from ncbi is actually sra file format. We can use fastq_dump program in SRA_toolkit to convert sra to fastq. http://www.ncbi.nlm.nih.gov/Traces/sra/?view=software
~/Downloads/sratoolkit.2.3.2-ubuntu64/bin/fastq-dump ~/Downloads/SRR034580.sra
If we want to run Galaxy locally, we can install it simply by 2 command lines
hg clone https://bitbucket.org/galaxy/galaxy-dist/ cd galaxy-dist hg update stable
To run Galaxy locally, we do
cd galaxy-dist sh run.sh --reload
The command line will show Starting server in PID XXXX. serving on http://127.0.0.1:8080. We can use Ctrl + C to stop the Galaxy process.
Note: One problem with this simple instruction is we have not created a user yet.
- Upload one fastq data. Click 'refresh' icon on the history panel to ensure the data is uploaded. Don't use 'refresh' button on the browser; it will cause an error for the current process.
- FASTQ Groomer. Convert the data to Galaxy needs. NGS: QC and manipulation => Illumina FASTQ. FASTQ quality scores type: Sanger. (~10 minutes). This part uses CPU not memory.
- Open a new browser tab and go to ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_6.1/all.dir/. Right click the file all.cDNA and copy link location. In Galaxy click 'Upload File from your computer' paste URL to URL/Text entry.
- Scroll down Galaxy and select NGS:Mapping -> Map with BWA. PS. for some reason, BWA is not available. So I use Bowtie2 instead. The output of Bowtie2 is bam file.
- For reference genome, choose 'Use one from the history'. Galaxy automatically find the reference file 'ftp://ftp.plantbiology....' from history.
- Library mate-paired => Single-end.
- FASTQ file => 2: FASTQ Groomer on data 1.
- BWA settings to use => Commonly Used.
- Execute (~ 15 minutes)
- We can view the alignment file (sam format) created from BWA by using UCSV or IGV (input is bam or bai format). We now use NGS: SAM Tools to convert sam file to bam file. Click 'SAM-to-BAM converts SAM format to BAM format' tool.
- Choose the source for the reference list => History
- Converts SAM file => 4: Map with BWA on data 2 and data 3.
- Using reference file => 3:ftp://ftp.plantbiology.....
- Execute (~5 minutes)
- We want to create bai file which is a shortcut to IGV. It breaks the data into smaller accessible chunks. So when you pull up a certain cDNA, it goes straight to the subset. Go to the history, click the pencil icon (Edit Attributes) on the file SAM-to-BAM on data 3 and data 4.
- Look at 'Convert to new format' section. Go ahead and click 'Convert'. (< 1 minute). This will create another file.
- Use browser and go to ftp website to download all.cDNA file to desktop. The desktop should contain 3 files - all.cDNA, rice.bam and rice.bai files for IGV.
- Goto http://www.broadinstitute.org/software/igv/download to download IGV which is a java-based application. I need to install java machine first by install openjdk-7-jdk. IGV by default will launch 'Human hg18' genome. Launch IGV by cd IGV_2.2.13;java -Xmx750m -jar igv.jar. I found the IGV input requires sam+bai OR bam+bai. So we need to click the pencil icon to create bai file first before we want to upload sam or bam file to IGV.
- Goto File => Import Genome. Call it 'rice' and select 'all.cDNA' sequence file. Click 'Save' button.
- Goto File => Upload from File => rice.bam.
- Top right panel is cDNA
- Middle right panel has a lot of 'boxes' which is a read. If we zoom in, we can see some read points to left (backward) while some points to right (forward). On the top is a histogram. For example, a base may be covered by a lot of reads then the histogram will show the high frequence.
- If we keep zoom in, we can see color at the Bottom right panel. Keeping zoom in, we can see the base G, C, T, A themselves.
- Using IGV, we can 1. examine coverage.
- We can 2. check 'alternative splicing'. (not for this cDNA)
- We can 3. examine SNPs for base change. If we see gray color (dark gray is hight quality read, light gray means low quality read), it means they are perfect match. If we see color, it means there is a change. For example, a read is 'C' but in fact it should be 'A'. If a case has many high quality reads, and half of them are 'G' but the reference genome shows 'A'. This is most likely a SNP. This is heterogeisity.
- Tophat - align RNA seq data to genomic DNA
- Suppose we have use Galaxy to upload 2 data. One is SRR034580 and we have run FASTQ Groomer on data 1. The second data is SRR034584 and we also have run FASTQ Groomer on data 2. We also have uploaded reference genome sequence.
- Goto Galaxy and find NGS: RNA Analysis => Tophat.
- reference genome => Use one from the history
- RNA-Seq FASTQ file => 2; FASTQ Groomer on data 1.
- Execute. This will create 2 files. One is splice junctions and the other is accepted_hits. We queue the job and run another Tophat with the 2nd 'groomer'ed data file. We are going to work on accepted_hits file.
- While the queue are running, we can click on 'pencil' icon on 'accepted_hits' job and run the utlity 'Convert to new format' (Bam to Bai). We should do this for both 'accepted_hits' files.
- For some reason, the execution failed: An error occurred with this dataset: TopHat v2.0.7 TopHat v2.0.7 Error indexing reference sequence /bin/sh: 1: bowtie-build: not found.
- Cufflinks. We will estimate transcript abundance by using FPKM (RPKM).
- SAM or BAM file of alignmed RNA-Seq reads => tophat on data 2.. accepted_hits
- Use Reference Annotation - No (choose Yes if we want annotation. This requires GTF format. See http://genome.ucsc.edu/FAQ/FAQformat.html#format4. We don't have it for rice.)
- Execute. This will create 3 files. Gene expression, transcript expression and assembled transcripts.
- We also run Cufflinks for 2nd accepted_hits file. (~ 25 minutes)
- Cuffcompare. Compare one to each other.
- GTF file produced by Cufflinks => assembled transcript from the 1st data
- Use another GTF file produced by Cufflinks => Yes. It automatically find the other one.
- Execute. (< 10 minutes). This will create 7 files. Transcript accuracy, tmap file & refmap flie from each assembled transcripts, combined transcripts and transcript tracking.
- We are interested in combined transcripts file (to use in Cuffdiff).
- Cuffdiff.
- Transcripts => combined transcripts.
- SAM or BAM file of aligned RNA-Seq reads => 1st accepted_hits
- SAM or BAM file or aligned RNA-Seq reads => 2nd accepted_hits
- Execute. This will generate 11 files. Isoform expression, gene expression, TSS groups expression, CDS Expression FPKM Tracking, isoform FPKM tracking, gene FPKM tracking, TSS groups FPKM tracking, CDS FPKM tracking, splicing diff, promoters diff, CDS diff. We are interested in 'gene expression' file. We can save it and open it in Excel.
- IGV - 2 RNA-Seq datasets aligned to genomic DNA using Tophat
- Load the reference genome rice (see above)
- Upload from file => rice4.bam. Upload from file => rice5.bam.
- Alternative RNA splicing.
edX course
PH525.5x Case Study: RNA-seq data analysis. The course notes are forming a book. Check out https://github.com/genomicsclass/labs and http://genomicsclass.github.io/book/.
Variant calling
Overview/papers
- Systematic comparison of variant calling pipelines using gold standard personal exome variants by Hwang 2015.
- A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data by Heng Li.
- http://www.slideshare.net/AustralianBioinformatics/introduction-to-nextgeneration
- http://www.slideshare.net/thomaskeane/overview-of-methods-for-variant-calling-from-nextgeneration-sequence-data-9608507
- http://www.slideshare.net/jandot/nextgeneration-sequencing-variation-discovery
Workflow
- Sequence reads -> Quality control -> Mapping -> Variant calling -> Filtering & Annotation -> Querying.
- Galaxy workflow guide for variant detection
- http://seqanswers.com/wiki/How-to/exome_analysis
- Alignment step includes
- mark PCR duplicates some reads will be exact copies of each other. These reads are called PCR duplicates due to amplification biases in PCR. They share the same sequence and the same alignment position and could cause trouble during SNP calling as possibly some allele is overrepresented due to amplification biases.
- local alignment around indels Indels within reads often lead to false positive SNPs at the end of sequence reads. To prevent this artifact, local realignment around indels is done using local realignment tools from the Genome Analysis Tool Kit.
- quality recalibration
- SNP calling step includes producing raw SNP calls and filter SNPs
- Annotation
- Alignment step includes
- WGS/WES Mapping to Variant Calls http://www.htslib.org/workflow/
Step 1: Mapping
bwa index <ref.fa> bwa mem -R '@RG\tID:foo\tSM:bar\tLB:library1' <ref.fa> <read1.fa> <read1.fa> > lane.sam samtools fixmate -O bam <lane.sam> <lane_fixmate.bam> samtools sort -O bam -o <lane_sorted.bam> -T </tmp/lane_temp> <lane_fixmate.sam>
Step 2: Improvement
java -Xmx2g -jar GenomeAnalysisTK.jar -T RealignerTargetCreator -R <ref.fa> -I <lane.bam> \
-o <lane.intervals> --known <bundle/b38/Mills1000G.b38.vcf>
java -Xmx4g -jar GenomeAnalysisTK.jar -T IndelRealigner -R <ref.fa> -I <lane.bam> \
-targetIntervals <lane.intervals> --known <bundle/b38/Mills1000G.b38.vcf> -o <lane_realigned.bam>
java -Xmx4g -jar GenomeAnalysisTK.jar -T BaseRecalibrator -R <ref.fa> \
-knownSites >bundle/b38/dbsnp_142.b38.vcf> -I <lane.bam> -o <lane_recal.table>
java -Xmx2g -jar GenomeAnalysisTK.jar -T PrintReads -R <ref.fa> -I <lane.bam> \
--BSQR <lane_recal.table> -o <lane_recal.bam>
java -Xmx2g -jar MarkDuplicates.jar VALIDATION_STRINGENCY=LENIENT \
INPUT=<lane_1.bam> INPUT=<lane_2.bam> INPUT=<lane_3.bam> OUTPUT=<library.bam>
samtools merge <sample.bam> <library1.bam> <library2.bam> <library3.bam>
samtools index <sample.bam>
Step 3: Variant calling
samtools mpileup -ugf <ref.fa> <sample1.bam> <sample2.bam> <sample3.bam> | \
bcftools call -vmO z -o <study.vcf.gz>
Non-R Software
Variant detector/discovery, genotyping
- Evaluation of variant detection software for pooled next-generation sequence data
- A survey of tools for variant analysis of next-generation genome sequencing data
- Evaluation of variant detection software for pooled next-generation sequence data
- Reliable Identification of Genomic Variants from RNA-Seq Data
- Bioinformatics at COMAV
Variant Identification
- Samtools
- https://wikis.utexas.edu/display/bioiteam/Variant+calling+using+SAMtools
- http://petridishtalk.com/2013/02/01/variant-discovery-annotation-filtering-with-samtools-the-gatk/
- http://lab.loman.net/2012/10/31/a-simple-guide-to-variant-calling-with-bwa-samtools-varscan2/
- Samtools and GATK
- Difference Between Samtools And Gatk Algorithms
- GATK by BROAD Institute.
- varscan2
- bcbio - Validated, scalable, community developed variant calling and RNA-seq analysis. Validated variant calling with human genome build 38.
- freebayes and a complete tutorial from alignment to variant calling.
GUI software
Variant Annotation
Biocoductor and R packages
- Bioconductor VariantTools package can export to VCF and VariantAnnotation package can read VCF format files. See also the Annotating Variants workflow in Bioconductor. See also SIFT.Hsapiens.dbSNP137, COSMIC.67, and PolyPhen.Hsapiens.dbSNP131 packages.
- R packages seqminer
fundamental - VariantAnnotation
readVcf() can be used to read a *.vcf or *.vcf.gz file.
dbSNP - SNPlocs.Hsapiens.dbSNP.20101109
Compare the rs numbers of the data and dbSNP.
variant wrt genes - TxDb.Hsapiens.UCSC.hg19.knownGene
Look for the column LOCATION which has values coding, fiveUTR, threeUTR, intron, intergenic, spliceSite, and promoter.
amino acid change for the non-synonymous variants - BSgenome.Hsapiens.UCSC.hg19
Look for the column CONSEQUENCE which has values synonymous or nonsynonymous.
SIFT & Polyphen for predict the impact of amino acid substitution on a human protein - PolyPhen.Hsapiens.dbSNP131
Look for the column PREDICTION which has values possibly damaging or benign.
vcf
vcf format
- One row is one snp.
- variant category (snp, insertion, deletion, mixed) http://www.1000genomes.org/node/101
- What is a VCF and how should I interpret it? from broad.
- See also bam format.
Open with LibreOffice Calc
Use delimiter 'Tab' only and uncheck the others.
It will then get the correct header #CHROM, POS, ID, REF, ALT, QUAL, FILTER, INFO (separated by ;), FORMAT (separated by :), SampleID (separated by :) where INFO field contains site-level annotations and FORMAT&SampleID fields contain sample-level annotations.
Count number of records, snps and indels
- vcftools.
- https://www.biostars.org/p/49386/, https://www.biostars.org/p/109086/, https://www.biostars.org/p/50923/, https://wikis.utexas.edu/display/bioiteam/Variant+calling+using+SAMtools.
- Summary statistics
bcftools stats XXX.vcf # SN, Summary numbers: # SN [2]id [3]key [4]value SN 0 number of samples: 0 SN 0 number of records: 71712 SN 0 number of no-ALTs: 0 SN 0 number of SNPs: 65950 SN 0 number of MNPs: 0 SN 0 number of indels: 5762 SN 0 number of others: 0 SN 0 number of multiallelic sites: 0 SN 0 number of multiallelic SNP sites: 0
- Count number of records
grep -c -v "^#" XXX.vcf # -c means count, -v is invert search
VariantsToTable tool from Braod GATK
VariantAnnotation package
- readVcf(), ScanVcfParam() and readInfo() functions.
- scanVcfHeader() and scanVcfParam() functions.
To install the R package, first we need to install required software in Linux
sudo apt-get update sudo apt-get install libxml2-dev sudo apt-get install libcurl4-openssl-dev
and then
source("http://bioconductor.org/biocLite.R")
biocLite("VariantAnnotation")
Example:
# The vcf file is created from running samtools on SRR1656687.fastq nskip=$(grep "^#" ~/Downloads/BMBC2_liver3_IMPACT_raw.vcf | wc -l) echo $nskip # 53 awk 'NR==53, NR==53' ~/Downloads/BMBC2_liver3_IMPACT_raw.vcf #CHROM POS ID REF ALT QUAL FILTER INFO FORMAT dedup.bam nall=$(cat ~/Downloads/BMBC2_liver3_IMPACT_raw.vcf | wc -l) echo $nall # 302174 # Generate <variantsOnly.vcf> which contains only the variant body awk 'NR==54, NR==302174' ~/Downloads/BMBC2_liver3_IMPACT_raw.vcf > variantsOnly.vcf # Generate <infoField.txt> which contains only the INFO field cut -f 8 variantsOnly.vcf > infoField.txt # Generate the final product head -n 1 infoField.txt DP=4;VDB=0.64;SGB=-0.453602;RPB=1;MQB=1;BQB=1;MQ0F=0;AC=2;AN=2;DP4=0,1,0,2;MQ=22 head -n 1 infoField.txt | cut -f1- -d ";" --output-delimiter " " ## Method 1: use cut ### Problem: the result is not a table since not all fields appear in all variants. cut -f1- -d ";" --output-delimiter=$'\t' infoField.txt > infoFieldTable.txt ## Method 2: use sed ### Problem: same as Method 1. sed 's/;/\t/g' infoField.txt > infoFieldTable2.txt ### OR if we want to replace the original file sed -i 's/;/\t/g' infoField.txt $ head -n 3 infoField.txt DP=4;VDB=0.64;SGB=-0.453602;RPB=1;MQB=1;BQB=1;MQ0F=0;AC=2;AN=2;DP4=0,1,0,2;MQ=22 DP=6;VDB=0.325692;SGB=-0.556411;MQ0F=0.333333;AC=2;AN=2;DP4=0,0,0,4;MQ=11 DP=5;VDB=0.0481133;SGB=-0.511536;RPB=1;MQB=1;MQSB=1;BQB=1;MQ0F=0;ICB=1;HOB=0.5;AC=1;AN=2;DP4=1,0,2,1;MQ=26 $ grep "^##INFO=<ID=" BMBC2_liver3_IMPACT_raw.vcf ##INFO=<ID=INDEL,Number=0,Type=Flag,Description="Indicates that the variant is an INDEL."> ##INFO=<ID=IDV,Number=1,Type=Integer,Description="Maximum number of reads supporting an indel"> ##INFO=<ID=IMF,Number=1,Type=Float,Description="Maximum fraction of reads supporting an indel"> ##INFO=<ID=DP,Number=1,Type=Integer,Description="Raw read depth"> ##INFO=<ID=VDB,Number=1,Type=Float,Description="Variant Distance Bias for filtering splice-site artefacts in RNA-seq data (bigger is better)",Version="3"> ##INFO=<ID=RPB,Number=1,Type=Float,Description="Mann-Whitney U test of Read Position Bias (bigger is better)"> ##INFO=<ID=MQB,Number=1,Type=Float,Description="Mann-Whitney U test of Mapping Quality Bias (bigger is better)"> ##INFO=<ID=BQB,Number=1,Type=Float,Description="Mann-Whitney U test of Base Quality Bias (bigger is better)"> ##INFO=<ID=MQSB,Number=1,Type=Float,Description="Mann-Whitney U test of Mapping Quality vs Strand Bias (bigger is better)"> ##INFO=<ID=SGB,Number=1,Type=Float,Description="Segregation based metric."> ##INFO=<ID=MQ0F,Number=1,Type=Float,Description="Fraction of MQ0 reads (smaller is better)"> ##INFO=<ID=ICB,Number=1,Type=Float,Description="Inbreeding Coefficient Binomial test (bigger is better)"> ##INFO=<ID=HOB,Number=1,Type=Float,Description="Bias in the number of HOMs number (smaller is better)"> ##INFO=<ID=AC,Number=A,Type=Integer,Description="Allele count in genotypes for each ALT allele, in the same order as listed"> ##INFO=<ID=AN,Number=1,Type=Integer,Description="Total number of alleles in called genotypes"> ##INFO=<ID=DP4,Number=4,Type=Integer,Description="Number of high-quality ref-forward , ref-reverse, alt-forward and alt-reverse bases"> ##INFO=<ID=MQ,Number=1,Type=Integer,Description="Average mapping quality"> odroid@odroid:~/Downloads$ $ head -n2 variantsOnly.vcf 1 10285 . T C 19.3175 . DP=4;VDB=0.64;SGB=-0.453602;RPB=1;MQB=1;BQB=1;MQ0F=0;AC=2;AN=2;DP4=0,1,0,2;MQ=22 GT:PL 1/1:46,2,0 1 10333 . C T 14.9851 . DP=6;VDB=0.325692;SGB=-0.556411;MQ0F=0.333333;AC=2;AN=2;DP4=0,0,0,4;MQ=11 GT:PL 1/1:42,12,0 $ awk 'NR==53, NR==58' ~/Downloads/BMBC2_liver3_IMPACT_raw.vcf #CHROM POS ID REF ALT QUAL FILTER INFO FORMAT dedup.bam 1 10285 . T C 19.3175 . DP=4;VDB=0.64;SGB=-0.453602;RPB=1;MQB=1;BQB=1;MQ0F=0;AC=2;AN=2;DP4=0,1,0,2;MQ=22 GT:PL 1/1:46,2,0 1 10333 . C T 14.9851 . DP=6;VDB=0.325692;SGB=-0.556411;MQ0F=0.333333;AC=2;AN=2;DP4=0,0,0,4;MQ=11 GT:PL 1/1:42,12,0 1 16257 . G C 24.566 . DP=5;VDB=0.0481133;SGB=-0.511536;RPB=1;MQB=1;MQSB=1;BQB=1;MQ0F=0;ICB=1;HOB=0.5;AC=1;AN=2;DP4=1,0,2,1;MQ=26 GT:PL 0/1:57,0,10 1 16534 . C T 13.9287 . DP=5;VDB=0.87268;SGB=-0.556411;MQ0F=0.4;AC=2;AN=2;DP4=0,0,4,0;MQ=11 GT:PL 1/1:41,12,0 1 16571 . G A 12.4197 . DP=7;VDB=0.560696;SGB=-0.556411;RPB=1;MQB=1;MQSB=0;BQB=1;MQ0F=0.714286;AC=2;AN=2;DP4=1,0,2,2;MQ=8 GT:PL 1/1:39,8,0 odroid@odroid:~/Downloads$
As you can see it is not easy to create a table from the INFO field.
Now we use the VariantAnnotation package.
> library("VariantAnnotation")
> v=readVcf("BMBC2_liver3_IMPACT_raw.vcf", "hg19")
> v
class: CollapsedVCF
dim: 302121 1
rowRanges(vcf):
GRanges with 5 metadata columns: paramRangeID, REF, ALT, QUAL, FILTER
info(vcf):
DataFrame with 17 columns: INDEL, IDV, IMF, DP, VDB, RPB, MQB, BQB, MQSB, ...
info(header(vcf)):
Number Type Description
INDEL 0 Flag Indicates that the variant is an INDEL.
IDV 1 Integer Maximum number of reads supporting an indel
IMF 1 Float Maximum fraction of reads supporting an indel
DP 1 Integer Raw read depth
VDB 1 Float Variant Distance Bias for filtering splice-site arte...
RPB 1 Float Mann-Whitney U test of Read Position Bias (bigger is...
MQB 1 Float Mann-Whitney U test of Mapping Quality Bias (bigger ...
BQB 1 Float Mann-Whitney U test of Base Quality Bias (bigger is ...
MQSB 1 Float Mann-Whitney U test of Mapping Quality vs Strand Bia...
SGB 1 Float Segregation based metric.
MQ0F 1 Float Fraction of MQ0 reads (smaller is better)
ICB 1 Float Inbreeding Coefficient Binomial test (bigger is better)
HOB 1 Float Bias in the number of HOMs number (smaller is better)
AC A Integer Allele count in genotypes for each ALT allele, in th...
AN 1 Integer Total number of alleles in called genotypes
DP4 4 Integer Number of high-quality ref-forward , ref-reverse, al...
MQ 1 Integer Average mapping quality
geno(vcf):
SimpleList of length 2: GT, PL
geno(header(vcf)):
Number Type Description
GT 1 String Genotype
PL G Integer List of Phred-scaled genotype likelihoods
>
> header(v)
class: VCFHeader
samples(1): dedup.bam
meta(2): META contig
fixed(2): FILTER ALT
info(17): INDEL IDV ... DP4 MQ
geno(2): GT PL
>
> dim(info(v))
[1] 302121 17
> info(v)[1:4, ]
DataFrame with 4 rows and 17 columns
INDEL IDV IMF DP VDB RPB
<logical> <integer> <numeric> <integer> <numeric> <numeric>
1:10285_T/C FALSE NA NA 4 0.6400000 1
1:10333_C/T FALSE NA NA 6 0.3256920 NA
1:16257_G/C FALSE NA NA 5 0.0481133 1
1:16534_C/T FALSE NA NA 5 0.8726800 NA
MQB BQB MQSB SGB MQ0F ICB
<numeric> <numeric> <numeric> <numeric> <numeric> <numeric>
1:10285_T/C 1 1 NA -0.453602 0.000000 NA
1:10333_C/T NA NA NA -0.556411 0.333333 NA
1:16257_G/C 1 1 1 -0.511536 0.000000 1
1:16534_C/T NA NA NA -0.556411 0.400000 NA
HOB AC AN DP4 MQ
<numeric> <IntegerList> <integer> <IntegerList> <integer>
1:10285_T/C NA 2 2 0,1,0,... 22
1:10333_C/T NA 2 2 0,0,0,... 11
1:16257_G/C 0.5 1 2 1,0,2,... 26
1:16534_C/T NA 2 2 0,0,4,... 11
> write.table(info(v)[1:4,], file="infoFieldTable3.txt", sep="\t", quote=F)
> head(rowRanges(v), 3)
GRanges object with 3 ranges and 5 metadata columns:
seqnames ranges strand | paramRangeID REF
<Rle> <IRanges> <Rle> | <factor> <DNAStringSet>
1:10285_T/C 1 [10285, 10285] * | <NA> T
1:10333_C/T 1 [10333, 10333] * | <NA> C
1:16257_G/C 1 [16257, 16257] * | <NA> G
ALT QUAL FILTER
<DNAStringSetList> <numeric> <character>
1:10285_T/C C 19.3175 .
1:10333_C/T T 14.9851 .
1:16257_G/C C 24.5660 .
-------
seqinfo: 25 sequences from hg19 genome
> qual(v)[1:5]
[1] 19.3175 14.9851 24.5660 13.9287 12.4197
> alt(v)[1:5]
DNAStringSetList of length 5
[[1]] C
[[2]] T
[[3]] C
[[4]] T
[[5]] A
> q('no')
vcfR package
Filtering strategy
- https://support.bioconductor.org/p/62817/ using QUAL and DP.
SAMtools (samtools, bcftools, htslib)
- http://samtools.sourceforge.net/mpileup.shtml
- http://massgenomics.org/2012/03/5-things-to-know-about-samtools-mpileup.html. Li's paper is available in NCBI PMC.
- A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data by Heng Li.
- https://www.broadinstitute.org/gatk/media/docs/Samtools.pdf
export bdge_bowtie_PATH=/opt/RNA-Seq/bin/bowtie2-2.2.1
export bdge_tophat_PATH=/opt/RNA-Seq/bin/tophat-2.0.11.Linux_x86_64
export bdge_samtools_PATH=/opt/RNA-Seq/bin/samtools-0.1.19:/opt/RNA-Seq/bin/samtools-0.1.19/bcftools:/opt/RNA-Seq/bin/samtools-0.1.19/misc
export PATH=$bdge_bowtie_PATH:$bdge_samtools_PATH:$bdge_tophat_PATH:$PATH
samtools sort TNBC1/accepted_hits.bam TNBC1/accepted_hits-sorted
samtools index TNBC1/accepted_hits-sorted.bam TNBC1/accepted_hits-sorted.bai
samtools mpileup -uf ~/igenome/human/NCBI/build37.2/genome.fa \
TNBC1/accepted_hits-sorted.bam | bcftools view -vcg - > TNBC1/var.raw.vcf
where '-u' in mpileup means uncompressed and '-f' means faidx indexed reference sequence file. If we do not use pipe command, the output from samtools mpileup is a bcf file which can be viewed by using 'bcftools view XXX.bcf | more' command.
Note that the samtools mpileup can be used in different ways. For example,
samtools mpileup -f XXX.fa XXX.bam > XXX.mpileup samtools mpileup -v -u -f XXX.fa XXX.bam > XXX.vcf samtools mpileup -g -f XXX.fa XXX.bam > XXX.bcf
bcftools
bcftools — utilities for variant calling and manipulating VCFs and their binary counterparts BCFs. bcftools is one of components in SAMtools software.
Installation
wget https://github.com/samtools/bcftools/releases/download/1.2/bcftools-1.2.tar.bz2 sudo tar jxf bcftools-1.2.tar.bz2 -C /opt/RNA-Seq/bin/ cd /opt/RNA-Seq/bin/bcftools-1.2/ sudo make # create bcftools, plot-vcfstats, vcfutils.pl commands
Example
samtools sort TNBC1/accepted_hits.bam TNBC1/accepted_hits-sorted
samtools index TNBC1/accepted_hits-sorted.bam TNBC1/accepted_hits-sorted.bai
samtools mpileup -uf ~/igenome/human/NCBI/build37.2/genome.fa \
TNBC1/accepted_hits-sorted.bam | bcftools view -vcg - > TNBC1/var.raw.vcf
# Or
samtools mpileup -g -f XXX.fa XXX.bam > sample.bcf
bcftools call -v -m -O z -o var.raw.vcf.gz sample.bcf
zcat var.raw.vcf.gz | more
zcat var.raw.vcf.gz | grep -v "^#" | wc -l
samtools tview -p 17:1234567 XXX.bam XXX.fa | more # IGV alternative
where '-v' means exports variants only, '-m' for multiallelic-caller and '-O z' means compressed vcf format.
To count the number of snps, indels, et al in the vcf file, use
bcftools stats xxx.vcf | more
htslib
- bgzip – Block compression/decompression utility. The output file .gz is in a binary format.
- tabix – Generic indexer for TAB-delimited genome position files. The output file tbi is in a binary format.
Installation
wget https://github.com/samtools/htslib/releases/download/1.2.1/htslib-1.2.1.tar.bz2 sudo tar jxf htslib-1.2.1.tar.bz2 -C /opt/RNA-Seq/bin/ cd /opt/RNA-Seq/bin/htslib-1.2.1/ sudo make # create tabix, htsfile, bgzip commands
Example
export PATH=/opt/RNA-Seq/bin/bcftools-1.2/:$PATH export PATH=/opt/RNA-Seq/bin/htslib-1.2.1/:$PATH # zip and index bgzip -c var.raw.vcf > var.raw.vcf.gz tabix var.raw.vcf.gz # create var.raw.vcf.ga.tbi, index vcf files (very fast in this step) bcftools annotate -c ID -a common_all_20150603.vcf.gz var.raw.vcf.gz > var_annot.vcf # 2 min # subset based on chromosome 1, include 'chr' and position range if necessary tabix -h var.raw.vcf.gz 1: > chr1.vcf tabix -h var.raw.vcf.gz chr1:10,000,000-20,000,000
GATK
- Source code is at github
- Need to register and create an account w/ password in order to download the software (require Java).
- GATK guide and requirements.
- Best Practices workflow for SNP and indel calling on RNAseq data using GATK. It recommend using HaplotypeCaller.
- Indel realignment Input=BAM, Output=BAM. Big improvement for Base Quality Score Recalibration when run on realigned BAM files (artificial SNPs are replaced with real indels).
- Base Quality Score Recalibration Input=BAM, known sites, Output=BAM.
- UnifiedGenotyper for variant call.
- https://wikis.utexas.edu/display/bioiteam/Variant+calling+with+GATK
- Identification of somatic and germline mutations using whole exome sequencing of congenital acute lymphoblastic leukemia: a paper that has used GATK for germline and somatic analyses
- http://gatkforums.broadinstitute.org/discussion/3892/the-gatk-best-practices-for-variant-calling-on-rnaseq-in-full-detail
- New plugin to support your BWA-GATK pipelines for Biomedical Genomics Server Solution
# Download GenomeAnalysisTK-3.4-46.tar.bz2 from gatk website sudo mkdir /opt/RNA-Seq/bin/gatk sudo tar jxvf ~/Downloads/GenomeAnalysisTK-3.4-46.tar.bz2 -C /opt/RNA-Seq/bin/gatk ls /opt/RNA-Seq/bin/gatk # GenomeAnalysisTK.jar resources
freebayes
The program gave different errors for some real datasets downloaded from GEO.
A small test data included in the software works.
brb@brb-P45T-A:~/github$ sudo apt-get update brb@brb-P45T-A:~/github$ sudo apt-get install cmake brb@brb-P45T-A:~/github$ git clone --recursive git://github.com/ekg/freebayes.git brb@brb-P45T-A:~/github$ cd freebayes/ brb@brb-P45T-A:~/github/freebayes$ make brb@brb-P45T-A:~/github/freebayes$ make test # Got some errors. So don't worry to 'make test' brb@brb-P45T-A:~/github/freebayes/test/tiny$ ../../bin/freebayes -f q.fa NA12878.chr22.tiny.bam > tmp.vcf brb@brb-P45T-A:~/github/freebayes/test/tiny$ ls -lt total 360 -rw-rw-r-- 1 brb brb 15812 Oct 9 10:52 tmp.vcf -rw-rw-r-- 1 brb brb 287213 Oct 9 09:33 NA12878.chr22.tiny.bam -rw-rw-r-- 1 brb brb 96 Oct 9 09:33 NA12878.chr22.tiny.bam.bai -rw-rw-r-- 1 brb brb 16307 Oct 9 09:33 NA12878.chr22.tiny.giab.vcf -rw-rw-r-- 1 brb brb 12565 Oct 9 09:33 q.fa -rw-rw-r-- 1 brb brb 16 Oct 9 09:33 q.fa.fai -rw-rw-r-- 1 brb brb 2378 Oct 9 09:33 q_spiked.vcf.gz -rw-rw-r-- 1 brb brb 91 Oct 9 09:33 q_spiked.vcf.gz.tbi -rw-rw-r-- 1 brb brb 4305 Oct 9 09:33 q.vcf.gz -rw-rw-r-- 1 brb brb 102 Oct 9 09:33 q.vcf.gz.tbi brb@brb-P45T-A:~/github/freebayes/test/tiny$ ~/github/samtools/samtools view NA12878.chr22.tiny.bam | wc -l 3333 brb@brb-P45T-A:~/github/freebayes/test/tiny$ wc -l tmp.vcf 76 tmp.vcf brb@brb-P45T-A:~/github/freebayes/test/tiny$ wc -l q.fa 207 q.fa
To debug the code, we can
../../bin/freebayes -f q.fa NA12878.chr22.tiny.bam > tmpFull.out 2>&1
Then compare tmp.vcf and tmpFull.out files. We see
- total sites: 12280
- the first variant call happens at position 186. Look at tmpFull.out file, we see most of positions just show 3 lines in the output but variants like position 186 show a lot of output (line 643 to 736).
- the output 'processing position XXX' was generated from AlleleParser:toNextPosition() which was called by AlleleParser::getNextAlleles() which was called by freebayes.cpp::main() line 126, the while() loop.
Varscan2
vcftools
https://vcftools.github.io/examples.html
This toolset can be used to perform the following operations on VCF files:
- Filter out specific variants
- Compare files
- Summarize variants
- Convert to different file types
- Validate and merge files
- Create intersections and subsets of variants
wget http://sourceforge.net/projects/vcftools/files/vcftools_0.1.12b.tar.gz/download \
-o vcftools_0.1.12b.tar.gz
tar -xzvf vcftools_0.1.12b.tar.gz
sudo mv vcftools_0.1.12b /opt/RNA-Seq/bin/
export PERL5LIB=/opt/RNA-Seq/bin/vcftools_0.1.12b/perl/
/opt/RNA-Seq/bin/vcftools_0.1.12b/
make
export PATH=$PATH:/opt/RNA-Seq/bin/vcftools_0.1.12b/bin
ls bin
# fill-aa vcf-annotate vcf-convert vcf-phased-join vcf-subset
# fill-an-ac vcf-compare vcf-fix-ploidy vcf-query vcftools
# fill-fs vcf-concat vcf-indel-stats vcf-shuffle-cols vcf-to-tab
# fill-ref-md5 vcf-consensus vcf-isec vcf-sort vcf-tstv
# man1 vcf-contrast vcf-merge vcf-stats vcf-validator
Some example
$ cd ~/SRP032789
$ vcftools --vcf GSM1261016_IP2-50_var.flt.vcf
VCFtools - v0.1.12b
(C) Adam Auton and Anthony Marcketta 2009
Parameters as interpreted:
--vcf GSM1261016_IP2-50_var.flt.vcf
After filtering, kept 1 out of 1 Individuals
After filtering, kept 193609 out of a possible 193609 Sites
Run Time = 1.00 seconds
$ wc -l GSM1261016_IP2-50_var.flt.vcf
193636 GSM1261016_IP2-50_var.flt.vcf
$ vcf-indel-stats < GSM1261016_IP2-50_var.flt.vcf > out.txt
Use of uninitialized value in pattern match (m//) at /opt/RNA-Seq/bin/vcftools_0.1.12b/bin/vcf-indel-stats line 49.
Use of uninitialized value in concatenation (.) or string at /opt/RNA-Seq/bin/vcftools_0.1.12b/bin/vcf-indel-stats line 49.
<: No such file or directory at /opt/RNA-Seq/bin/vcftools_0.1.12b/bin/vcf-indel-stats line 18.
main::error('<: No such file or directory') called at /opt/RNA-Seq/bin/vcftools_0.1.12b/bin/vcf-indel-stats line 50
main::init_regions('HASH(0xd77cb8)') called at /opt/RNA-Seq/bin/vcftools_0.1.12b/bin/vcf-indel-stats line 71
main::do_stats('HASH(0xd77cb8)') called at /opt/RNA-Seq/bin/vcftools_0.1.12b/bin/vcf-indel-stats line 9
To compare two vcf files, see https://vcftools.github.io/documentation.html
./vcftools --vcf input_data.vcf --diff other_data.vcf --out compare
Online course on Variant calling
- edX: HarvardX: PH525.6x Case Study: Variant Discovery and Genotyping. Course notes is at their Github page.
Variant Annotation
See also the paper A survey of tools for variant analysis of next-generation genome sequencing data.
dbSNP
SNPlocs data R package for Human. Some clarification about SNPlocs.Hsapiens.dbSNP.20120608 package.
> library(BSgenome) > available.SNPs() [1] "SNPlocs.Hsapiens.dbSNP141.GRCh38" [2] "SNPlocs.Hsapiens.dbSNP142.GRCh37" [3] "SNPlocs.Hsapiens.dbSNP.20090506" [4] "SNPlocs.Hsapiens.dbSNP.20100427" [5] "SNPlocs.Hsapiens.dbSNP.20101109" [6] "SNPlocs.Hsapiens.dbSNP.20110815" [7] "SNPlocs.Hsapiens.dbSNP.20111119" [8] "SNPlocs.Hsapiens.dbSNP.20120608" [9] "XtraSNPlocs.Hsapiens.dbSNP141.GRCh38"
An example:
wget https://github.com/samtools/bcftools/releases/download/1.2/bcftools-1.2.tar.bz2 sudo tar jxf bcftools-1.2.tar.bz2 -C /opt/RNA-Seq/bin/ cd /opt/RNA-Seq/bin/bcftools-1.2/ sudo make wget https://github.com/samtools/htslib/releases/download/1.2.1/htslib-1.2.1.tar.bz2 sudo tar jxf htslib-1.2.1.tar.bz2 -C /opt/RNA-Seq/bin/ cd /opt/RNA-Seq/bin/htslib-1.2.1/ sudo make # create tabix, htsfile, bgzip commands export bdge_bowtie_PATH=/opt/RNA-Seq/bin/bowtie2-2.2.1 export bdge_tophat_PATH=/opt/RNA-Seq/bin/tophat-2.0.11.Linux_x86_64 export bdge_samtools_PATH=/opt/RNA-Seq/bin/samtools-0.1.19 export PATH=$bdge_bowtie_PATH:$bdge_samtools_PATH:$bdge_tophat_PATH:$PATH export PATH=/opt/RNA-Seq/bin/bcftools-1.2/:$PATH export PATH=/opt/RNA-Seq/bin/htslib-1.2.1/:$PATH wget ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606/VCF/common_all_20150603.vcf.gz.tbi wget ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606/VCF/common_all_20150603.vcf.gz cd TNBC1 mv ~/Downloads/common_all_20150603.vcf.gz* . bgzip -c var.raw.vcf > var.raw.vcf.gz tabix var.raw.vcf.gz bcftools annotate -c ID -a common_all_20150603.vcf.gz var.raw.vcf.gz > var_annot.vcf
Any found in dbSNP?
grep -c 'rs[0-9]' raw_snps.vcf
ANNOVAR
ANNOVAR and the web based interface to ANNOVAR wANNOVAR. ANNOVAR can annotate genetic variants using
- Gene-based annotation
- Region-based annotation
- Filter-based annotation
- Other functionalities
Note that
- It is correct to use the original download link from the email to download the latest version.
- annovar folder needs to be placed under a directory with write permission
- If we run annovar with a new reference genome, the code will need to download some database. When annovar is downloading the database, the cpu is resting so the whole process looks idling.
- Annovar will print out a warning with information about changes if there is a new version available.
- BRB-SeqTools (grep "\.pl" preprocessgui/*.*) uses annotate_variation.pl (5), convert2annovar.pl (2), table_annovar.pl (4), variants_reduction.pl (1).
- In nih/biowulf, it creates $ANNOVAR_HOME and $ANNOVAR_DATA environment variables.
- To check the version of my local copy
~/annovar/annotate_variation.pl
- Contents of annovar
brb@T3600 ~ $ ls -l ~/annovar/ total 476 -rwxr-xr-x 1 brb brb 212090 Mar 7 14:59 annotate_variation.pl -rwxr-xr-x 1 brb brb 13589 Mar 7 14:59 coding_change.pl -rwxr-xr-x 1 brb brb 166582 Mar 7 14:59 convert2annovar.pl drwxr-xr-x 2 brb brb 4096 Jun 18 2015 example drwxr-xr-x 3 brb brb 4096 Mar 21 09:59 humandb -rwxr-xr-x 1 brb brb 19419 Mar 7 14:59 retrieve_seq_from_fasta.pl -rwxr-xr-x 1 brb brb 34682 Mar 7 14:59 table_annovar.pl -rwxr-xr-x 1 brb brb 21774 Mar 7 14:59 variants_reduction.pl brb@T3600 ~ $ ls -lh ~/annovar/humandb | head total 36G -rw-r--r-- 1 brb brb 927 Mar 21 09:59 annovar_downdb.log drwxr-xr-x 2 brb brb 4.0K Jun 18 2015 genometrax-sample-files-gff -rw-r--r-- 1 brb brb 20K Mar 7 14:59 GRCh37_MT_ensGeneMrna.fa -rw-r--r-- 1 brb brb 3.1K Mar 7 14:59 GRCh37_MT_ensGene.txt -rw-r--r-- 1 brb brb 1.4G Dec 15 2014 hg19_AFR.sites.2014_10.txt -rw-r--r-- 1 brb brb 87M Dec 15 2014 hg19_AFR.sites.2014_10.txt.idx -rw-r--r-- 1 brb brb 2.8G Dec 15 2014 hg19_ALL.sites.2014_10.txt -rw-r--r-- 1 brb brb 89M Dec 15 2014 hg19_ALL.sites.2014_10.txt.idx -rw-r--r-- 1 brb brb 978M Dec 15 2014 hg19_AMR.sites.2014_10.txt
SnpEff & SnpSift
- http://snpeff.sourceforge.net/SnpEff.html (basic information)
- http://snpeff.sourceforge.net/SnpEff_manual.html (Manual)
- http://snpeff.sourceforge.net/protocol.html (6 examples)
- https://wiki.hpcc.msu.edu/display/Bioinfo/SnpEff+-+The+Basics
- https://docs.uabgrid.uab.edu/wiki/Galaxy_DNA-Seq_Tutorial
- http://www.genome.gov/Pages/Research/DIR/DIRNewsFeatures/Next-Gen101/Teer_VariantAnnotation.pdf
- http://veda.cs.uiuc.edu/course2013/pres/fields/lab/Variant_Calling_v1.pptx (if you click the link, chrome will dl the pptx file)
- In BRB-SeqTools, snpEff.jar (2) and SnpSift.jar (5)
- NIH/biowulf also has installed snpEff & SnpSift
SnpEff: Genetic variant annotation and effect prediction toolbox
- Input: vcf & reference genome database (eg GRCh38.79).
- Output: vcf & <snpEff_summary.html> & < snpEff_genes.txt> files.
wget http://iweb.dl.sourceforge.net/project/snpeff/snpEff_latest_core.zip
sudo unzip snpEff_latest_core.zip -d /opt/RNA-Seq/bin
export PATH=/opt/RNA-Seq/bin/snpEff/:$PATH
# Next we want to download snpEff database.
# 1. Need to pay attention the database is snpEff version dependent
# 2. Instead of using the command line (very slow < 1MB/s),
# java -jar /opt/RNA-Seq/bin/snpEff/snpEff.jar databases | grep GRCh
# java -jar /opt/RNA-Seq/bin/snpEff/snpEff.jar download GRCh38.79
# we just go to the file using the the web browser
# http://sourceforge.net/projects/snpeff/files/databases/v4_1/ # 525MB
# 3. the top folder of the zip file is called 'data'. We will need to unzip it to the snpEff directory.
# That is, snpEff +- data
# |- examples
# |- galaxy
# +- scripts
mv ~/Downloads/snpEff_v4_1_GRCh38.79.zip .
unzip snpEff_v4_1GRCh38.79.zip
sudo java -Xmx4G -jar /opt/RNA-Seq/bin/snpEff/snpEff.jar \
-i vcf -o vcf GRCh38.79 var_annot.vcf > var_annot_snpEff.vcf
brb@T3600 ~ $ ls -l /opt/SeqTools/bin/snpEff/
total 44504
drwxrwxr-x 5 brb brb 4096 May 5 11:24 data
drwxr-xr-x 2 brb brb 4096 Feb 17 16:37 examples
drwxr-xr-x 3 brb brb 4096 Feb 17 16:37 galaxy
drwxr-xr-x 3 brb brb 4096 Feb 17 16:37 scripts
-rw-r--r-- 1 brb brb 6138594 Dec 5 10:49 snpEff.config
-rw-r--r-- 1 brb brb 20698856 Dec 5 10:49 snpEff.jar
-rw-r--r-- 1 brb brb 18712032 Dec 5 10:49 SnpSift.jar
Output file (compare cosmic_dbsnp_rem.vcf and snpeff_anno.vcf at here):
- add 5 lines at the header. Search "ID=ANN" at snpeff_anno.vcf
##SnpEffVersion="4.2 (build 2015-12-05), by Pablo Cingolani" ##SnpEffCmd="SnpEff -no-downstream -no-upstream .... ##INFO=<ID=ANN,Number=.,Type=String,Description="Functional annotations: 'Allele | Annotation | Annotation_Impact | Gene_Name | Gene_ID | Feature_Type | Feature_ID |... ##INFO=<ID=LOF,Number=.,Type=String,Description="Predicted loss of function effects for this variant. ... ##INFO=<ID=NMD,Number=.,Type=String,Description="Predicted nonsense mediated decay effects for this variant. ...
- added functional annotations ANN in the INFO field. See an example at Basic example: Annotate using SnpEff
ANN=A|missense_variant|MODERATE|ISG15|ISG15|transcript|NM_005101.3|protein_coding|2/2|c.248G>A|p.Ser83Asn|355/666|248/498|83/165||
SnpSift: SnpSift helps filtering and manipulating genomic annotated files
Once you annotated your files using SnpEff, you can use SnpSift to help you filter large genomic datasets in order to find the most significant variants
cat "$outputDir/tmp/$tmpfd/snpeff_anno.vcf" | \
java -jar "$seqtools_snpeff/SnpSift.jar" \
filter "(ANN[*].BIOTYPE = 'protein_coding') | (ANN[*].EFFECT has 'splice')" \
> "$outputDir/tmp/$tmpfd/snpeff_proteincoding.vcf"
the output file (compare snpeff_anno.vcf and snpeff_proteincoding.vcf at https://github.com/arraytools/vc-annotation/tree/master/snpeff/tmp here]
- the header will add 3 lines
##SnpSiftVersion="SnpSift 4.2 (build 2015-12-05), by Pablo Cingolani" ##SnpSiftCmd="SnpSift filter '(ANN[*].BIOTYPE = 'protein_coding') | (ANN[*].EFFECT has 'splice')'" ##FILTER=<ID=SnpSift,Description="SnpSift 4.2 ...
- KEEP variants satisfying the filter criterion (In the ANN field, BIOTYPE=protein_coding and EFFECT=splice). So it will reduce the variants size. No more fields are added.
java -jar "$seqtools_snpeff/SnpSift.jar" dbNSFP -f $dbnsfpField \
-v -db "$dbnsfpFile" "$outputDir/tmp/$tmpfd/nonsyn_splicing2.vcf" \
> "$outputDir/tmp/$tmpfd/nonsyn_splicing_dbnsfp.vcf"
Compare nonsyn_splicing2.vcf and nonsyn_splicing_dbnsfp.vcf at github output file
- the header adds 29 lines.
##SnpSiftCmd="SnpSift dbnsfp -f SIFT_score,SIFT_pred, ... ##INFO=<ID=dbNSFP_GERP___RS,Number=A,Type=Float,Description="Field 'GERP++_RS' from dbNSFP"> ##INFO=<ID=dbNSFP_CADD_phred,Number=A,Type=Float,Description="Field 'CADD_phred' from dbNSFP"> ...
- the INFO field will add
dbNSFP_CADD_phred=2.276;dbNSFP_CADD_raw=-0.033441;dbNSFP_FATHMM_pred=.,.,T; ...
java -jar "$seqtools_snpeff/SnpSift.jar" extractFields \
-s "," -e "." "$outputDir/tmp/$tmpfd/nonsyn_splicing_dbnsfp.vcf" CHROM POS ID REF ALT "ANN[0].EFFECT" "ANN[0].IMPACT" "ANN[0].GENE" "ANN[0].GENEID" "ANN[0].FEATURE" "ANN[0].FEATUREID" "ANN[0].BIOTYPE" "ANN[0].HGVS_C" "ANN[0].HGVS_P" dbNSFP_SIFT_score dbNSFP_SIFT_pred dbNSFP_Polyphen2_HDIV_score dbNSFP_Polyphen2_HDIV_pred dbNSFP_Polyphen2_HVAR_score dbNSFP_Polyphen2_HVAR_pred dbNSFP_LRT_score dbNSFP_LRT_pred dbNSFP_MutationTaster_score dbNSFP_MutationTaster_pred dbNSFP_MutationAssessor_score dbNSFP_MutationAssessor_pred dbNSFP_FATHMM_score dbNSFP_FATHMM_pred dbNSFP_PROVEAN_score dbNSFP_PROVEAN_pred dbNSFP_VEST3_score dbNSFP_CADD_raw dbNSFP_CADD_phred dbNSFP_MetaSVM_score dbNSFP_MetaSVM_pred dbNSFP_MetaLR_score dbNSFP_MetaLR_pred dbNSFP_GERP___NR dbNSFP_GERP___RS dbNSFP_phyloP100way_vertebrate dbNSFP_phastCons100way_vertebrate dbNSFP_SiPhy_29way_logOdds \
> "$outputDir/tmp/$tmpfd/annoTable.txt"
The output file will
- remove header from the input VCF file
- break the INFO field into several columns; Only the columns we specify in the command will be kept.
Local
$ ls -l /opt/SeqTools/bin/snpEff/ total 44504 drwxrwxr-x 5 brb brb 4096 May 5 11:24 data drwxr-xr-x 2 brb brb 4096 Feb 17 16:37 examples drwxr-xr-x 3 brb brb 4096 Feb 17 16:37 galaxy drwxr-xr-x 3 brb brb 4096 Feb 17 16:37 scripts -rw-r--r-- 1 brb brb 6138594 Dec 5 10:49 snpEff.config -rw-r--r-- 1 brb brb 20698856 Dec 5 10:49 snpEff.jar -rw-r--r-- 1 brb brb 18712032 Dec 5 10:49 SnpSift.jar $ ls -l /opt/SeqTools/bin/snpEff/data total 12 drwxr-xr-x 2 brb brb 4096 May 5 11:24 GRCh38.82 drwxrwxr-x 2 brb brb 4096 Feb 5 09:28 hg19 drwxr-xr-x 2 brb brb 4096 Mar 8 09:44 hg38
Biowulf
The following code fixed some typos on biowulf website.
echo $SNPSIFT_JAR
# Make sure the database (GRCH37.75 in this case) exists
ls /usr/local/apps/snpEff/4.2/data
# CanFam3.1.75 Felis_catus_6.2.75 GRCh37.75 GRCh38.82 GRCm38.81 GRCz10.82 hg38
# CanFam3.1.81 Felis_catus_6.2.81 GRCh37.GTEX GRCh38.p2.RefSeq GRCm38.82 hg19 hg38kg
# CanFam3.1.82 Felis_catus_6.2.82 GRCh38.81 GRCm38.75 GRCz10.81 hg19kg Zv9.75
# Use snpEff to annotate against GRCh37.75
snpEff -v -lof -motif -hgvs -nextProt GRCh37.75 protocols/ex1.vcf > ex1.eff.vcf # 25 minutes
# create ex1.eff.vcf (475MB), snpEff_genes.txt (2.5MB) and snpEff_summary.html (22MB)
# Use SnpSift to pull out 'HIGH IMPACT' or 'MODERATE IMPACT' variants
cat ex1.eff.vcf | \
java -jar $SNPSIFT_JAR filter "((EFF[*].IMPACT = 'HIGH') | (EFF[*].IMPACT = 'MODERATE'))" \
> ex1.filtered.vcf
# ex1.filtered.vcf (8.2MB), 2 minutes
# Use SnpSift to annotate against the dbNSFP database
java -jar $SNPSIFT_JAR dbnsfp -v -db /fdb/dbNSFP2/dbNSFP2.9.txt.gz ex1.eff.vcf \
> file.annotated.vcf
# file.annotated.vcf (479 MB), 11 minutes
and the output
$ ls -lth | head total 994M -rw-r----- 1 479M May 15 11:47 file.annotated.vcf -rw-r----- 1 8.2M May 15 11:31 ex1.filtered.vcf -rw-r----- 1 476M May 15 10:31 ex1.eff.vcf -rw-r----- 1 2.5M May 15 10:30 snpEff_genes.txt -rw-r----- 1 22M May 15 10:30 snpEff_summary.html lrwxrwxrwx 1 39 May 15 10:01 protocols -> /usr/local/apps/snpEff/4.2/../protocols
COSMIC
- Youtube videos
- Histogram of BRAF (melanoma).
AnnTools
GNS-SNP
SeattleSeq
SVA
VARIANT
VEP
Web tools
- UCSC Variant Annotation Integrator
- SeattleSeq Annotation 137
- Variant Effect Predictor and VEP script
De novo genome assembly
Single Cell RNA-Seq
monocle
sincell
SCell
SCell – integrated analysis of single-cell RNA-seq data
The best RNA-Seq analysis interface
Systematically evaluating interfaces for RNA-seq analysis from a life scientist perspective
Co-expression in RNA-Seq
Monitor Software Version Change
Circos Plot
Circos is a popular tool for summarizing genomic events in a tumor genome.
Cancer & Mutation
My Cancer Genome
CIViC - Clinical Interpretations of Variants in Cancer
NCBI
- NCBI Resources and Variant Interpretation Tools for the Clinical Community: ClinVar, MedGen, GTR, Variation Viewer
R and Bioconductor packages
- Some basics of biomaRt (and GenomicRanges)
- Annotating Ranges Represent common sequence data types (e.g., from BAM, gff, bed, and wig files) as genomic ranges for simple and advanced range-based queries.
library(VariantAnnotation) library(AnnotationHub) library(TxDb.Hsapiens.UCSC.hg19.knownGene) library(TxDb.Mmusculus.UCSC.mm10.ensGene) library(org.Hs.eg.db) library(org.Mm.eg.db) library(BSgenome.Hsapiens.UCSC.hg19)
- Analysis of RNA-Seq Data with R/Bioconductor by Girke in UC Riverside
- R / Bioconductor for High-Throughput Sequence Analysis 2012 by Martin Morgan1 and Nicolas Delhomme
- Count-based differential expression analysis of RNA sequencing data using R and Bioconductor by Simon Anders
- Sequences, Genomes, and Genes in R / Bioconductor by Martin Morgan 2013.
- Orchestrating high-throughput genomic analysis with Bioconductor by Wolfgang Huber et al 2015.
- RNA-seq Analysis Example This is a script that will do differential gene expression (DGE) analysis for RNA-seq experiments using the bioconductor package edgeR. RPKMs were calculated for bar plots.
Some workflows
RNA-Seq workflow
Gene-level exploratory analysis and differential expression. A non stranded-specific and paired-end rna-seq experiment was used for the tutorial.
STAR Samtools Rsamtools
fastq -----> sam ----------> bam ----------> bamfiles -|
\ GenomicAlignments DESeq2
--------------------> se --------> dds
GenomicFeatures GenomicFeatures / (SummarizedExperiment) (DESeqDataSet)
gtf ----------------> txdb ---------------> genes -----|
Sequence analysis
library(ShortRead) or library(Biostrings) (QA) gtf + library(GenomicFeatures) or directly library(TxDb.Scerevisiae.UCSC.sacCer2.sgdGene) (gene information) GenomicRanges::summarizeOverlaps or GenomicRanges::countOverlaps(count) edgeR or DESeq2 (gene expression analysis) library(org.Sc.sgd.db) or library(biomaRt)
Accessing Annotation Data
Use microarray probe, gene, pathway, gene ontology, homology and other annotations. Access GO, KEGG, NCBI, Biomart, UCSC, vendor, and other sources.
library(org.Hs.eg.db) # Sample OrgDb Workflow
library("hgu95av2.db") # Sample ChipDb Workflow
library(TxDb.Hsapiens.UCSC.hg19.knownGene) # Sample TxDb Workflow
library(Homo.sapiens) # Sample OrganismDb Workflow
library(AnnotationHub) # Sample AnnotationHub Workflow
library("biomaRt") # Using biomaRt
library(BSgenome.Hsapiens.UCSC.hg19) # BSgenome packages
| Object type | example package name | contents |
|---|---|---|
| OrgDb | org.Hs.eg.db | gene based information for Homo sapiens |
| TxDb | TxDb.Hsapiens.UCSC.hg19.knownGene | transcriptome ranges for Homo sapiens |
| OrganismDb | Homo.sapiens | composite information for Homo sapiens |
| BSgenome | BSgenome.Hsapiens.UCSC.hg19 | genome sequence for Homo sapiens |
| refGenome |
RNA-Seq Data Analysis using R/Bioconductor
- https://github.com/datacarpentry/rnaseq-data-analysis by Stephen Turner.
- Tutorial: Introduction to Bioconductor for high-throughput sequence analysis by UseR 2015
- systemPipeR Building end-to-end analysis pipelines with automated report generation for next generation sequence (NGS) applications such as RNA-Seq, ChIP-Seq, VAR-Seq and Ribo-Seq. An important feature is support for running command-line software, such as NGS aligners, on both single machines or compute clusters.
- BioC2015
limma
- Differential expression analyses for RNA-sequencing and microarray studies
- Case Study using a Bioconductor R pipeline to analyze RNA-seq data (this is linked from limma package user guide). Here we illustrate how to use two Bioconductor packages - Rsubread' and limma - to perform a complete RNA-seq analysis, including Subread'Bold text read mapping, featureCounts read summarization, voom normalization and limma differential expresssion analysis.
- Unbalanced data, non-normal data, Bartlett's test for equal variance across groups and SAM tests (assumes equal variances just like limma). See this post.
easyRNASeq
Calculates the coverage of high-throughput short-reads against a genome of reference and summarizes it per feature of interest (e.g. exon, gene, transcript). The data can be normalized as 'RPKM' or by the 'DESeq' or 'edgeR' package.
ShortRead
Base classes, functions, and methods for representation of high-throughput, short-read sequencing data.
Rsamtools
The Rsamtools package provides an interface to BAM files.
The main purpose of the Rsamtools package is to import BAM files into R. Rsamtools also provides some facility for file access such as record counting, index file creation, and filtering to create new files containing subsets of the original. An important use case for Rsamtools is as a starting point for creating R objects suitable for a diversity of work flows, e.g., AlignedRead objects in the ShortRead package (for quality assessment and read manipulation), or GAlignments objects in GenomicAlignments package (for RNA-seq and other applications). Those desiring more functionality are encouraged to explore samtools and related software efforts
This package provides an interface to the 'samtools', 'bcftools', and 'tabix' utilities (see 'LICENCE') for manipulating SAM (Sequence Alignment / Map), FASTA, binary variant call (BCF) and compressed indexed tab-delimited (tabix) files.
IRanges
IRanges is a fundamental package (see how many packages depend on it) to other packages like GenomicRanges, GenomicFeatures and GenomicAlignments. The package defines the IRanges class.
The plotRanges() function given in the 'An Introduction to IRanges' vignette shows how to draw an IRanges object.
If we want to make the same plot using the ggplot2 package, we can follow the example in this post. Note that disjointBins() returns a vector the bin number for each bins counting on the y-axis.
flank
The example is obtained from ?IRanges::flank.
ir3 <- IRanges(c(2,5,1), c(3,7,3)) # IRanges of length 3 # start end width # [1] 2 3 2 # [2] 5 7 3 # [3] 1 3 3 flank(ir3, 2) # start end width # [1] 0 1 2 # [2] 3 4 2 # [3] -1 0 2 # Note: by default flank(ir3, 2) = flank(ir3, 2, start = TRUE, both=FALSE) # For example, [2,3] => [2,X] => (..., 0, 1, 2) => [0, 1] # == == flank(ir3, 2, start=FALSE) # start end width # [1] 4 5 2 # [2] 8 9 2 # [3] 4 5 2 # For example, [2,3] => [X,3] => (..., 3, 4, 5) => [4,5] # == == flank(ir3, 2, start=c(FALSE, TRUE, FALSE)) # start end width # [1] 4 5 2 # [2] 3 4 2 # [3] 4 5 2 # Combine the ideas of the previous 2 cases. flank(ir3, c(2, -2, 2)) # start end width # [1] 0 1 2 # [2] 5 6 2 # [3] -1 0 2 # The original statement is the same as flank(ir3, c(2, -2, 2), start=T, both=F) # For example, [5, 7] => [5, X] => ( 5, 6) => [5, 6] # == == flank(ir3, -2, start=F) # start end width # [1] 2 3 2 # [2] 6 7 2 # [3] 2 3 2 # For example, [5, 7] => [X, 7] => (..., 6, 7) => [6, 7] # == == flank(ir3, 2, both = TRUE) # start end width # [1] 0 3 4 # [2] 3 6 4 # [3] -1 2 4 # The original statement is equivalent to flank(ir3, 2, start=T, both=T) # (From the manual) If both = TRUE, extends the flanking region width positions into the range. # The resulting range thus straddles the end point, with width positions on either side. # For example, [2, 3] => [2, X] => (..., 0, 1, 2, 3) => [0, 3] # == # == == == == flank(ir3, 2, start=FALSE, both=TRUE) # start end width # [1] 2 5 4 # [2] 6 9 4 # [3] 2 5 4 # For example, [2, 3] => [X, 3] => (..., 2, 3, 4, 5) => [4, 5] # == # == == == ==
Both IRanges and GenomicRanges packages provide the flank function.
Flanking region is also a common term in High-throughput sequencing. The IGV user guide also has some option related to flanking.
- General tab: Feature flanking regions (base pairs). IGV adds the flank before and after a feature locus when you zoom to a feature, or when you view gene/loci lists in multiple panels.
- Alignments tab: Splice junction track options. The minimum amount of nucleotide coverage required on both sides of a junction for a read to be associated with the junction. This affects the coverage of displayed junctions, and the display of junctions covered only by reads with small flanking regions.
GenomicRanges
GenomicRanges depends on IRanges package. See the dependency diagram below.
GenomicFeatues ------- GenomicRanges -+- IRanges -- BioGenomics
| +
+-----+ +- GenomeInfoDb
| |
GenomicAlignments +--- Rsamtools --+-----+
+--- Biostrings
The package defines some classes
- GRanges
- GRangesList
- GAlignments
- SummarizedExperiment: it has the following slots - expData, rowData, colData, and assays. Accessors include assays(), assay(), colData(), expData(), mcols(), ... The mcols() method is defined in the S4Vectors package.
(As of Jan 6, 2015) The introduction in GenomicRanges vignette mentions the GAlignments object created from a 'BAM' file discarding some information such as SEQ field, QNAME field, QUAL, MAPQ and any other information that is not needed in its document. This means that multi-reads don't receive any special treatment. Also pair-end reads will be treated as single-end reads and the pairing information will be lost. This might change in the future.
GenomicAlignments
Counting reads with summarizeOverlaps vignette
library(GenomicAlignments)
library(DESeq)
library(edgeR)
fls <- list.files(system.file("extdata", package="GenomicAlignments"),
recursive=TRUE, pattern="*bam$", full=TRUE)
features <- GRanges(
seqnames = c(rep("chr2L", 4), rep("chr2R", 5), rep("chr3L", 2)),
ranges = IRanges(c(1000, 3000, 4000, 7000, 2000, 3000, 3600, 4000,
7500, 5000, 5400), width=c(rep(500, 3), 600, 900, 500, 300, 900,
300, 500, 500)), "-",
group_id=c(rep("A", 4), rep("B", 5), rep("C", 2)))
features
# GRanges object with 11 ranges and 1 metadata column:
# seqnames ranges strand | group_id
# <Rle> <IRanges> <Rle> | <character>
# [1] chr2L [1000, 1499] - | A
# [2] chr2L [3000, 3499] - | A
# [3] chr2L [4000, 4499] - | A
# [4] chr2L [7000, 7599] - | A
# [5] chr2R [2000, 2899] - | B
# ... ... ... ... ... ...
# [7] chr2R [3600, 3899] - | B
# [8] chr2R [4000, 4899] - | B
# [9] chr2R [7500, 7799] - | B
# [10] chr3L [5000, 5499] - | C
# [11] chr3L [5400, 5899] - | C
# -------
# seqinfo: 3 sequences from an unspecified genome; no seqlengths
olap
# class: SummarizedExperiment
# dim: 11 2
# exptData(0):
# assays(1): counts
# rownames: NULL
# rowData metadata column names(1): group_id
# colnames(2): sm_treated1.bam sm_untreated1.bam
# colData names(0):
assays(olap)$counts
# sm_treated1.bam sm_untreated1.bam
# [1,] 0 0
# [2,] 0 0
# [3,] 0 0
# [4,] 0 0
# [5,] 5 1
# [6,] 5 0
# [7,] 2 0
# [8,] 376 104
# [9,] 0 0
# [10,] 0 0
# [11,] 0 0
Pasilla data. Note that the bam files are not clear where to find them. According to the message, we can download SAM files first and then convert them to BAM files by samtools (Not verify yet).
samtools view -h -o outputFile.bam inputFile.sam
A modified R code that works is
###################################################
### code chunk number 11: gff (eval = FALSE)
###################################################
library(rtracklayer)
fl <- paste0("ftp://ftp.ensembl.org/pub/release-62/",
"gtf/drosophila_melanogaster/",
"Drosophila_melanogaster.BDGP5.25.62.gtf.gz")
gffFile <- file.path(tempdir(), basename(fl))
download.file(fl, gffFile)
gff0 <- import(gffFile, asRangedData=FALSE)
###################################################
### code chunk number 12: gff_parse (eval = FALSE)
###################################################
idx <- mcols(gff0)$source == "protein_coding" &
mcols(gff0)$type == "exon" &
seqnames(gff0) == "4"
gff <- gff0[idx]
## adjust seqnames to match Bam files
seqlevels(gff) <- paste("chr", seqlevels(gff), sep="")
chr4genes <- split(gff, mcols(gff)$gene_id)
###################################################
### code chunk number 12: gff_parse (eval = FALSE)
###################################################
library(GenomicAlignments)
# fls <- c("untreated1_chr4.bam", "untreated3_chr4.bam")
fls <- list.files(system.file("extdata", package="pasillaBamSubset"),
recursive=TRUE, pattern="*bam$", full=TRUE)
path <- system.file("extdata", package="pasillaBamSubset")
bamlst <- BamFileList(fls)
genehits <- summarizeOverlaps(chr4genes, bamlst, mode="Union") # SummarizedExperiment object
assays(genehits)$counts
###################################################
### code chunk number 15: pasilla_exoncountset (eval = FALSE)
###################################################
library(DESeq)
expdata = MIAME(
name="pasilla knockdown",
lab="Genetics and Developmental Biology, University of
Connecticut Health Center",
contact="Dr. Brenton Graveley",
title="modENCODE Drosophila pasilla RNA Binding Protein RNAi
knockdown RNA-Seq Studies",
pubMedIds="20921232",
url="http://www.ncbi.nlm.nih.gov/projects/geo/query/acc.cgi?acc=GSE18508",
abstract="RNA-seq of 3 biological replicates of from the Drosophila
melanogaster S2-DRSC cells that have been RNAi depleted of mRNAs
encoding pasilla, a mRNA binding protein and 4 biological replicates
of the the untreated cell line.")
design <- data.frame(
condition=c("untreated", "untreated"),
replicate=c(1,1),
type=rep("single-read", 2), stringsAsFactors=TRUE)
library(DESeq)
geneCDS <- newCountDataSet(
countData=assay(genehits),
conditions=design)
experimentData(geneCDS) <- expdata
sampleNames(geneCDS) = colnames(genehits)
###################################################
### code chunk number 16: pasilla_genes (eval = FALSE)
###################################################
chr4tx <- split(gff, mcols(gff)$transcript_id)
txhits <- summarizeOverlaps(chr4tx, bamlst)
txCDS <- newCountDataSet(assay(txhits), design)
experimentData(txCDS) <- expdata
We can also check out ?summarizeOverlaps to find some fake examples.
Rsubread
See this post for about C version of the featureCounts program.
Inference
DESeq or edgeR
- DESeq2 method
- DESeq2 with a large number of samples -> use DESEq2 to normalize the data and then use do a Wilcoxon rank-sum test on the normalized counts, for each gene separately, or, even better, use a permutation test. See this post. Or consider the limma-voom method instead, which will handle 1000 samples in a few seconds without the need for extra memory.
- edgeR normalization factor post. Normalization factors are computed using the trimmed mean of M-values (TMM) method; see the paper by Robinson & Oshlack 2010 for more details. Briefly, M-values are defined as the library size-adjusted log-ratio of counts between two libraries. The most extreme 30% of M-values are trimmed away, and the mean of the remaining M-values is computed. This trimmed mean represents the log-normalization factor between the two libraries. The idea is to eliminate systematic differences in the counts between libraries, by assuming that most genes are not DE.
- Can I feed TCGA normalized count data to EdgeR?
- counts() function and normalized counts.
- Why use Negative binomial distribution in RNA-Seq data?] and the Presentation by Simon Anders.
prebs
Probe region expression estimation for RNA-seq data for improved microarray comparability
DEXSeq
Inference of differential exon usage in RNA-Seq
rSeqNP
A non-parametric approach for detecting differential expression and splicing from RNA-Seq data
Pipeline
SARTools
pasilla and pasillaBamSubset Data
pasilla - Data package with per-exon and per-gene read counts of RNA-seq samples of Pasilla knock-down by Brooks et al., Genome Research 2011.
pasillaBamSubset - Subset of BAM files untreated1.bam (single-end reads) and untreated3.bam (paired-end reads) from "Pasilla" experiment (Pasilla knock-down by Brooks et al., Genome Research 2011).
BitSeq
Transcript expression inference and differential expression analysis for RNA-seq data. The homepage of Antti Honkela.
ReportingTools
The ReportingTools software package enables users to easily display reports of analysis results generated from sources such as microarray and sequencing data.
sequences
More or less an educational package. It has 2 c and c++ source code. It is used in Advanced R programming and package development.
QuasR
Bioinformatics paper
CRAN packages
ssizeRNA
Sample Size Calculation for RNA-Seq Experimental Design
rbamtools
Provides an interface to functions of the 'SAMtools' C-Library by Heng Li
refGenome
The packge contains functionality for import and managing of downloaded genome annotation Data from Ensembl genome browser (European Bioinformatics Institute) and from UCSC genome browser (University of California, Santa Cruz) and annotation routines for genomic positions and splice site positions.
WhopGenome
Provides very fast access to whole genome, population scale variation data from VCF files and sequence data from FASTA-formatted files. It also reads in alignments from FASTA, Phylip, MAF and other file formats. Provides easy-to-use interfaces to genome annotation from UCSC and Bioconductor and gene ontology data from AmiGO and is capable to read, modify and write PLINK .PED-format pedigree files.
TCGA2STAT
Simple TCGA Data Access for Integrated Statistical Analysis in R
TCGA2STAT depends on Bioconductor package CNTools which cannot be installed automatically.
source("https://bioconductor.org/biocLite.R")
biocLite("CNTools")
install.packages("TCGA2STAT")
The getTCGA() function allows to download various kind of data:
- gene expression which includes mRNA-microarray gene expression data (data.type="mRNA_Array") & RNA-Seq gene expression data (data.type="RNASeq")
- miRNA expression which includes miRNA-array data (data.type="miRNA_Array") & miRNA-Seq data (data.type="miRNASeq")
- mutation data (data.type="Mutation")
- methylation expression (data.type="Methylation")
- copy number changes (data.type="CNA_SNP")
caOmicsV
http://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-016-0989-6 Data from TCGA ws used
Visualize multi-dimentional cancer genomics data including of patient information, gene expressions, DNA methylations, DNA copy number variations, and SNP/mutations in matrix layout or network layout.
Map2NCBI
The GetGeneList() function is useful to download Genomic Features (including gene features/symbols) from NCBI (ftp://ftp.ncbi.nih.gov/genomes/MapView/).
> library(Map2NCBI)
> GeneList = GetGeneList("Homo sapiens", build="ANNOTATION_RELEASE.107", savefiles=TRUE, destfile=path.expand("~/"))
# choose [2], [n], and [1] to filter the build and feature information.
# The destination folder will contain seq_gene.txt, seq_gene.md.gz and GeneList.txt files.
> str(GeneList)
'data.frame': 52157 obs. of 15 variables:
$ tax_id : chr "9606" "9606" "9606" "9606" ...
$ chromosome : chr "1" "1" "1" "1" ...
$ chr_start : num 11874 14362 17369 30366 34611 ...
$ chr_stop : num 14409 29370 17436 30503 36081 ...
$ chr_orient : chr "+" "-" "-" "+" ...
$ contig : chr "NT_077402.3" "NT_077402.3" "NT_077402.3" "NT_077402.3" ...
$ ctg_start : num 1874 4362 7369 20366 24611 ...
$ ctg_stop : num 4409 19370 7436 20503 26081 ...
$ ctg_orient : chr "+" "-" "-" "+" ...
$ feature_name : chr "DDX11L1" "WASH7P" "MIR6859-1" "MIR1302-2" ...
$ feature_id : chr "GeneID:100287102" "GeneID:653635" "GeneID:102466751" "GeneID:100302278" ...
$ feature_type : chr "GENE" "GENE" "GENE" "GENE" ...
$ group_label : chr "GRCh38.p2-Primary" "GRCh38.p2-Primary" "GRCh38.p2-Primary" "GRCh38.p2-Primary" ...
$ transcript : chr "Assembly" "Assembly" "Assembly" "Assembly" ...
$ evidence_code: chr "-" "-" "-" "-" ...
> GeneList$feature_name[grep("^NAP", GeneList$feature_name)]
Simulate RNA-Seq
http://en.wikipedia.org/wiki/List_of_RNA-Seq_bioinformatics_tools#RNA-Seq_simulators
BEERS/Grant G.R. 2011
http://bioinformatics.oxfordjournals.org/content/27/18/2518.long#sec-2. The simulation method is called BEERS and it was used in the STAR software paper.
sudo apt-get install cpanminus sudo cpanm Math::Random wget http://cbil.upenn.edu/BEERS/beers.tar wget http://itmat.rum.s3.amazonaws.com/simulator_config_human.tar.gz # 1.2GB tar -xvf beers.tar # two perl files perl reads_simulator.pl # Help
SimSeq
A data-based simulation algorithm for rna-seq data. The vector of read counts simulated for a given experimental unit has a joint distribution that closely matches the distribution of a source rna-seq dataset provided by the user.
empiricalFDR.DESeq2
http://biorxiv.org/content/early/2014/12/05/012211
The key function is simulateCounts, which takes a fitted DESeq2 data object as an input and returns a simulated data object (DESeq2 class) with the same sample size factors, total counts and dispersions for each gene as in real data, but without the effect of predictor variables.
Functions fdrTable, fdrBiCurve and empiricalFDR compare the DESeq2 results obtained for the real and simulated data, compute the empirical false discovery rate (the ratio of the number of differentially expressed genes detected in the simulated data and their number in the real data) and plot the results.
polyester
http://biorxiv.org/content/early/2014/12/05/012211
Given a set of annotated transcripts, polyester will simulate the steps of an RNA-seq experiment (fragmentation, reverse-complementing, and sequencing) and produce files containing simulated RNA-seq reads.
Input: reference FASTA file (containing names and sequences of transcripts from which reads should be simulated) OR a GTF file denoting transcript structures, along with one FASTA file of the DNA sequence for each chromosome in the GTF file.
Output: FASTA files. Reads in the FASTA file will be labeled with the transcript from which they were simulated.
RSEM
GEO
See the internal link at R-GEO.
DNA Seq Data
NIH
- Go to SRA/Sequence Read Archiveand type the keywords 'Whole Genome Sequencing human'. An example of the procedures to search whole genome sequencing data from human samples:
- Enter 'Whole Genome Sequencing human' in ncbi/sra search sra objects at http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=search_obj
- The webpage will return the result in terms of SRA experiments, SRA studies, Biosamples, GEO datasets. I pick SRA studies from Public Access.
- The result is sorted by the Accession number (does not take the first 3 letters like DRP into account). The Accession number has a format SRPxxxx. So I just go to the Last page (page 98)
- I pick the first one Accession:SRP066837 from this page. The page shows the Study type is whole genome sequence. http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?study=SRP066837
- (Important trick) Click the number next to Run. It will show a summary (SRR #, library name, MBases, age, biomaterial provider, isolate and sex) about all samples. http://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP066837
- Download the raw data from any one of them (eg SRR2968056). For whole genome, the Strategy is WGS. For whole exome, the Strategy is called WXS.
- Search the keywords 'nonsynonymous' and 'human' in PMC
Use SRATool Kit instead of wget to download
Don't use the wget command since it requires the specification of right http address.
Downloading SRA data using command line utilities
(Method 1) Use the fastq-dump command. For example, the following command (modified from the document will download the first 5 reads and save it to a file called <SRR390728.fastq> (NOT sra format) in the current directory.
/opt/RNA-Seq/bin/sratoolkit.2.3.5-2-ubuntu64/bin/fastq-dump -X 5 SRR390728 -O . # OR /opt/RNA-Seq/bin/sratoolkit.2.3.5-2-ubuntu64/bin/fastq-dump --split-3 SRR390728 # no progress bar
This will download the files in FASTQ format.
(Method 2) If we need to downloading by wget or FTP (works for ‘SRR’, ‘ERR’, or ‘DRR’ series):
wget ftp://ftp-trace.ncbi.nih.gov/sra/sra-instant/reads/ByRun/sra/SRR/SRR304/SRR304976/SRR304976.sra
It will download the file in SRA format. In the case of SRR590795, the sra is 240M and fastq files are 615*2MB.
(Method 3) Download Ubuntu x86_64 tarball from http://downloads.asperasoft.com/en/downloads/8?list
brb@T3600 ~/Downloads $ tar xzvf aspera-connect-3.6.2.117442-linux-64.tar.gz aspera-connect-3.6.2.117442-linux-64.sh brb@T3600 ~/Downloads $ ./aspera-connect-3.6.2.117442-linux-64.sh Installing Aspera Connect Deploying Aspera Connect (/home/brb/.aspera/connect) for the current user only. Restart firefox manually to load the Aspera Connect plug-in Install complete. brb@T3600 ~/Downloads $ ~/.aspera/connect/bin/ascp -QT -l640M \ -i ~/.aspera/connect/etc/asperaweb_id_dsa.openssh \ [email protected]:/sra/sra-instant/reads/ByRun/sra/SRR/SRR590/SRR590795/SRR590795.sra . SRR590795.sra 100% 239MB 535Mb/s 00:06 Completed: 245535K bytes transferred in 7 seconds (272848K bits/sec), in 1 file. brb@T3600 ~/Downloads $
Aspera is typically 10 times faster than FTP according to the website. For this case, wget takes 12s while ascp uses 7s.
Note that the URL on the website's is wrong. I got the correct URL from emailing to ncbi help. Google: ascp "[email protected]"
SRAdb package
https://bioconductor.org/packages/release/bioc/html/SRAdb.html
First we install some required package for XML and RCurl.
sudo apt-get update sudo apt-get install libxml2-dev sudo apt-get install libcurl4-openssl-dev
and then
source("https://bioconductor.org/biocLite.R")
biocLite("SRAdb")
SRA
Only the cancer types with expected cases > 10^5 in the US in 2015 are considered here. http://www.cancer.gov/types/common-cancers
SRP066363 - lung cancer
- http://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP066363
- http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE74866
SRP015769 or SRP062882 - prostate cancer
- http://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP015769 5 are from normal and 5 are from tumor. Whole Exome Seq.
- http://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP062882 6 normal and the rest are tumor.
SRP053134 - breast cancer
Look at the MBases value column. It determines the coverage for each run.
SRP050992 single cell RNA-Seq
Used in Design and computational analysis of single-cell RNA-sequencing experiments
Single cell RNA-Seq
Exploiting single-cell expression to characterize co-expression replicability
SRP040626 or SRP040540 - Colon and rectal cancer
- http://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP040626
- http://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP040540
Tutorials
See the BWA section.
Whole Exome Seq
- 1000genomes. 1000genomes and tcga are two places to get vcf files too.
- Review of Current Methods, Applications, and Data Management for the Bioinformatics Analysis of Whole Exome Sequencing (Bao 2014)
- A framework for variation discovery and genotyping using next-generation DNA sequencing data. See the table 1 there.
- Some data from SRA repository.
SraRunTable.txt
- http://www.ncbi.nlm.nih.gov/sra/?term=SRA059511
- http://www.ncbi.nlm.nih.gov/sra/SRX194938[accn] and click SRP004077
- http://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP004077 and click Runs from the RHS
- http://trace.ncbi.nlm.nih.gov/Traces/study/?acc=SRP004077 and click RunInfoTable
Note that (For this study, it has 2377 rows)
- Column A (AssemblyName_s) eg GRCh37
- Column I (library_name_s) eg
- column N (header=Run_s) shows all SRR or ERR accession numbers.
- Column P (Sample_Name)
- Column Y (header=Assay_Type_s) shows WGS.
- Column AB (LibraryLayout_s): PAIRED
Public Data
ISB Cancer Genomics Cloud (ISB-CGC)
http://cgc.systemsbiology.net/ Leveraging Google Cloud Platform for TCGA Analysis
NCI's Genomic Data Commons (GDC)
The GDC supports several cancer genome programs at the NCI Center for Cancer Genomics (CCG), including The Cancer Genome Atlas (TCGA), Therapeutically Applicable Research to Generate Effective Treatments (TARGET), and the Cancer Genome Characterization Initiative (CGCI).
Gene set analysis
Hypergeometric test
- A course from bioinformatics.ca and over-represented pathway.
- How informative are enrichment analyses really?
Misc
High performance
Merge different datasets (different genechips)
Normalization
- How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets
- Comparing the normalization methods for the differential analysis of Illumina high-throughput RNA-Seq data
How to use UCSC Table Browser
- An instruction from BitSeq software
Where to download reference genome
- UCSC and twoBitToFa to UCSC convert .2bit to fasta.
- hg19 from UCSC (chromosome-wise).
UCSC version & NCBI release corresponding
Gene Annotation
- GeneCards
- Genetics Home Reference from National Library of Medicine
- My Cancer Genome
- Cosmic
- annotables R package.
How many DNA strands are there in humans?
- http://www.numberof.net/number-of-dna-strands/
- http://www.answers.com/Q/How_many_DNA_strands_are_there_in_humans
How many base pairs in human
- chromosome 22 has the smallest number of bps (~50 million).
- chromosome 1 has the largest number of bps (245 base pairs).
NGS technology
DNA methylation
- GC content = (G+C)/(A+T+G+C) x 100%
- How many CpGs (C follows by G)?
- Analyzing DNA methylation data (part of the book Biomedical Data Science) and the PH525x: Data Analysis for Genomics (edX course). The Github website is on https://github.com/genomicsclass/labs. The source code may not be correct. See also http://www.biostat.jhsph.edu/~iruczins/teaching/kogo/html/ml/week8/methylation.Rmd. The paper Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies the tutorial has mentioned.
devtools::install_github("coloncancermeth","genomicsclass")
library(coloncancermeth) # 485512 x 26
data(coloncancermeth) # load meth (methylation data), pd (sample info ) and gr objects
dim(meth)
dim(pd)
length(gr)
colnames(pd)
table(pd$Status) # 9 normals, 17 cancers
normalIndex <- which(pd$Status=="normal")
cancerlIndex <- which(pd$Status=="cancer")
i=normalIndex[1]
plot(density(meth[,i],from=0,to=1),main="",ylim=c(0,3),type="n")
for(i in normalIndex){
lines(density(meth[,i],from=0,to=1),col=1)
}
### Add the cancer samples
for(i in cancerlIndex){
lines(density(meth[,i],from=0,to=1),col=2)
}
# finding regions of the genome that are different between cancer and normal samples
library(limma)
X<-model.matrix(~pd$Status)
fit<-lmFit(meth,X)
eb <- ebayes(fit)
# plot of the region surrounding the top hit
library(GenomicRanges)
i <- which.min(eb$p.value[,2])
middle <- gr[i,]
Index<-gr%over%(middle+10000)
cols=ifelse(pd$Status=="normal",1,2)
chr=as.factor(seqnames(gr))
pos=start(gr)
plot(pos[Index],fit$coef[Index,2],type="b",xlab="genomic location",ylab="difference")
matplot(pos[Index],meth[Index,],col=cols,xlab="genomic location")
# http://www.ncbi.nlm.nih.gov/pubmed/22422453
# within each chromosome we usually have big gaps creating subgroups of regions to be analyzed
chr1Index <- which(chr=="chr1")
hist(log10(diff(pos[chr1Index])),main="",xlab="log 10 method")
library(bumphunter)
cl=clusterMaker(chr,pos,maxGap=500)
table(table(cl)) ##shows the number of regions with 1,2,3, ... points in them
#consider two example regions#
...
Whole Genome Sequencing, Whole Exome Sequencing, Transcriptome (RNA) Sequencing
- http://www.rna-seqblog.com/exome-sequencing-vs-rna-seq-to-identify-coding-region-variants/
- http://www.rna-seqblog.com/combined-use-of-exome-and-transcriptome-sequencing/
- A comparison of whole genome and whole transcriptome sequencing
Sequence + Expression
RNASeq + ChipSeq
Labs
Biowulf2 at NIH
- Main site: http://hpc.nih.gov
- User guide: https://hpc.nih.gov/docs/user_guides.html
- Unlock account (60 days inactive) https://hpc.nih.gov/dashboard/
- Transitioning from PBS to Slurm: https://hpc.nih.gov/docs/pbs2slurm.html
- Job Submission 'cheat sheet': https://hpc.nih.gov/docs/biowulf2-handout.pdf
- STAR: https://hpc.nih.gov/apps/STAR.html
BamHash
Hash BAM and FASTQ files to verify data integrity. The C++ code is based on OpenSSL and seqan libraries.
Selected Papers
- Testing for association between RNA-Seq and high-dimensional data and the Bioconductor package globalSeq.
- The FDA’s Experience with Emerging Genomics Technologies—Past, Present, and Future
- Differential analysis of gene regulation at transcript resolution with RNA-seq Trapnell et al, Nature Biotechnology 31, 46–53 (2013)
Terms
RNA sequencing 101
Web
- Introduction to RNA-Seq including biology overview (DNA, Alternative splcing, mRNA structure, human genome) and sequencing technology.
- RNA Sequencing 101 by Agilent Technologies. Includes the definition of sequencing depth (number of reads per sample) and coverage (number of reads/locus).
- Where do we get reads(A,C,T,G) from sample RNA? See page 12 of this pdf from Colin Dewey in U. Wisc.
- Quantification of RNA-Seq data (see the above pdf)
- Convert read counts into expression: RPKM (see the above pdf)
- RPKM and FPKM (Data analysis of RNA-seq from new generation sequencing by 張庭毓) RNA-Seq資料分析研討會與實作課程 / RNA Seq定序 / 次世代定序(NGS) / 高通量基因定序 分析.
- First vs Second generation sequence.
- The Simple Fool’s Guide to Population Genomics via RNASeq: An Introduction to HighThroughput Sequencing Data Analysis. This covers QC, De novo assembly, BLAST, mapping reads to reference sequences, gene expression analysis and variant (SNP) detection.
- An Introduction to Bioinformatics Resources and their Practical Applications from NIH library Bioinformatics Support Program.
- Teaching material from rnaseqforthenextgeneration.org which includes Designing RNA-Seq experiments, Processing RNA-Seq data, and Downstream analyses with RNA-Seq data.
Books
- RNA-seq Data Analysis: A Practical Approach. The pdf version is available on slideshare.net.
- Statistical Analysis of Next Generation Sequencing Data
strand-specific vs non-strand specific experiment
- http://seqanswers.com/forums/showthread.php?t=28025. According to this message and the article (under the paragraph of Read counting) from PLOS, most of the RNA-seq protocols that are used nowadays are not strand-specific.
- http://biology.stackexchange.com/questions/1958/difference-between-strand-specific-and-not-strand-specific-rna-seq-data
- https://www.biostars.org/p/61625/ how to find if this public rnaseq data are prepared by strand-specific assay?
- https://www.biostars.org/p/62747/ Discussion of using IGV to view strand-specific coverage. See also the similar posts on the right hand side.
- https://www.biostars.org/p/44319/ How To Find Stranded Rna-Seq Experiments Data. The text dUTP 2nd strand marking includes a link to stranded rna-seq data.
- forward (+)/ reverse(-) strand in GAlignments objects (p68 of the pdf manual and page 7 of sam format specification.
Understand this info is necessary when we want to use summarizeOverlaps() function (GenomicAlignments) or htseq-count python program to get count data.
This post mentioned to use infer_experiment.py script to check whether the rna-seq run is stranded or not.
The rna-seq experiment used in this tutorial is not stranded-specific.
FASTQ
- FASTQ=FASTA + Qual. FASTQ format is a text-based format for storing both a biological sequence (usually nucleotide sequence) and its corresponding quality scores.
Phred quality score
q = -10log10(p) where p = error probability for the base.
| q | probability | base call accuracy |
|---|---|---|
| 10 | 0.1 | 90% |
| 20 | 0.01 | 99% |
| 30 | 0.001 | 99.9% |
| 40 | 0.0001 | 99.99% |
| 50 | 0.00001 | 99.999% |
FASTA <=> FASTQ conversion
Convert FASTA to FASTQ without quality scores
Biostars. For example, the bioawk by lh3 (Heng Li) worked.
Convert FASTA to FASTQ with quality score file
See the links on the above post.
Convert FASTQ to FASTA using Seqtk
Use the Seqtk program; see this post.
The Seqtk program by lh3 can be used to sample reads from a fastq file including paired-end; see this post.
RPKM (Mortazavi et al. 2008)
Reads per Kilobase of Exon per Million of Mapped reads.
- rpkm function in edgeR package.
- RPKM function in easyRNASeq package.
Idea
- The more we sequence, the more reads we expect from each gene. This is the most relevant correction of this method.
- Longer transcript are expected to generate more reads. The latter is only relevant for comparisons among different genes which we rarely perform!. As such, the DESeq2 only creates a size factor for each library and normalize the counts by dividing counts by a size factor (scalar) for each library. Note that: H0: mu1=mu2 is equivalent to H0: c*mu1=c*mu2 where c is gene length.
Calculation
- Count up the total reads in a sample and divide that number by 1,000,000 – this is our “per million” scaling factor.
- Divide the read counts by the “per million” scaling factor. This normalizes for sequencing depth, giving you reads per million (RPM)
- Divide the RPM values by the length of the gene, in kilobases. This gives you RPKM.
Formula
RPKM = (10^9 * C)/(N * L), with C = Number of reads mapped to a gene N = Total mapped reads in the experiment L = gene length in base-pairs for a gene
source("http://www.bioconductor.org/biocLite.R")
biocLite("edgeR")
library(edgeR)
set.seed(1234)
y <- matrix(rnbinom(20,size=1,mu=10),5,4)
[,1] [,2] [,3] [,4]
[1,] 0 0 5 0
[2,] 6 2 7 3
[3,] 5 13 7 2
[4,] 3 3 9 11
[5,] 1 2 1 15
d <- DGEList(counts=y, lib.size=1001:1004)
# Note that lib.size is optional
# By default, lib.size = colSums(counts)
cpm(d) # counts per million
Sample1 Sample2 Sample3 Sample4
1 0.000 0.000 4985.045 0.000
2 5994.006 1996.008 6979.063 2988.048
3 4995.005 12974.052 6979.063 1992.032
4 2997.003 2994.012 8973.081 10956.175
5 999.001 1996.008 997.009 14940.239
> cpm(d,log=TRUE)
Sample1 Sample2 Sample3 Sample4
1 7.961463 7.961463 12.35309 7.961463
2 12.607393 11.132027 12.81875 11.659911
3 12.355838 13.690089 12.81875 11.129470
4 11.663897 11.662567 13.17022 13.451207
5 10.285119 11.132027 10.28282 13.890078
d$genes$Length <- c(1000,2000,500,1500,3000)
rpkm(d)
Sample1 Sample2 Sample3 Sample4
1 0.0000 0.000 4985.0449 0.000
2 2997.0030 998.004 3489.5314 1494.024
3 9990.0100 25948.104 13958.1256 3984.064
4 1998.0020 1996.008 5982.0538 7304.117
5 333.0003 665.336 332.3363 4980.080
> cpm
function (x, ...)
UseMethod("cpm")
<environment: namespace:edgeR>
> showMethods("cpm")
Function "cpm":
<not an S4 generic function>
> cpm.default
function (x, lib.size = NULL, log = FALSE, prior.count = 0.25,
...)
{
x <- as.matrix(x)
if (is.null(lib.size))
lib.size <- colSums(x)
if (log) {
prior.count.scaled <- lib.size/mean(lib.size) * prior.count
lib.size <- lib.size + 2 * prior.count.scaled
}
lib.size <- 1e-06 * lib.size
if (log)
log2(t((t(x) + prior.count.scaled)/lib.size))
else t(t(x)/lib.size)
}
<environment: namespace:edgeR>
> rpkm.default
function (x, gene.length, lib.size = NULL, log = FALSE, prior.count = 0.25,
...)
{
y <- cpm.default(x = x, lib.size = lib.size, log = log, prior.count = prior.count)
gene.length.kb <- gene.length/1000
if (log)
y - log2(gene.length.kb)
else y/gene.length.kb
}
<environment: namespace:edgeR>
Here for example the 1st sample and the 2nd gene, its rpkm value is calculated as
# step 1: 6/(1.0e-6 *1001) = 5994.006 # cpm, compute column-wise # step 2: 5994.006/ (2000/1.0e3) = 2997.003 # rpkm, compute row-wise # Another way # step 1 (RPK) 6/ (2000/1.0e3) = 3 # step 2 (RPKM) 3/ (1.0e-6 * 1001) = 2997.003
Critics
Consider the following example: in two libraries, each with one million reads, gene X may have 10 reads for treatment A and 5 reads for treatment B, while it is 100x as many after sequencing 100 millions reads from each library. In the latter case we can be much more confident that there is a true difference between the two treatments than in the first one. However, the RPKM values would be the same for both scenarios. Thus, RPKM/FPKM are useful for reporting expression values, but not for statistical testing!
(another critic) Union Exon Based Approach
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0141910
In general, the methods for gene quantification can be largely divided into two categories: transcript-based approach and ‘union exon’-based approach.
It was found that the gene expression levels are significantly underestimated by ‘union exon’-based approach, and the average of RPKM from ‘union exons’-based method is less than 50% of the mean expression obtained from transcript-based approach.
FPKM (Trapnell et al. 2010)
Fragment per Kilobase of exon per Million of Mapped fragments (Cufflinks). FPKM is very similar to RPKM. RPKM was made for single-end RNA-seq, where every read corresponded to a single fragment that was sequenced. FPKM was made for paired-end RNA-seq. With paired-end RNA-seq, two reads can correspond to a single fragment, or, if one read in the pair did not map, one read can correspond to a single fragment. The only difference between RPKM and FPKM is that FPKM takes into account that two reads can map to one fragment (and so it doesn’t count this fragment twice).
RPKM, FPKM and TPM
- The youtube video is on here
- R code for computing effective counts, TPM, and FPKM
This is for the read normalization
P -- per K -- kilobase (related to gene length) M -- million (related to sequencing depth)
TMM (Robinson and Oshlack, 2010)
Trimmed Means of M values (EdgeR).
Coverage
- bedtools. The bedtools is now hosted on github
- https://github.com/alyssafrazee/polyester
~20x coverage ----> reads per transcript = transcriptlength/readlength * 20
- Page 18 of this RNA-Seq 101 from Agilent or Estimating Sequencing Coverage from Illumina.
C = L N / G
where L=read length, N =number of reads and G=haploid genome length. So, if we take one lane of single read human sequence with v3 chemistry, we get C = (100 bp)*(189×10^6)/(3×10^9 bp) = 6.3. This tells us that each base in the genome will be sequenced between six and seven times on average.
- coverage() function in IRanges package.
- Coverage and read depth
- What Is The Sequencing 'Depth' ? Coverage = (total number of bases generated) / (size of genome sequenced). So a 30x coverage means, on an average each base has been read by 30 sequences.
- Sequencing depth and coverage: key considerations in genomic analyses
- https://www.biostars.org/p/5165/
# Assume the bam file is sorted by chromosome location
# took 40 min on 5.8G bam file
samtools depth *bamfile* | awk '{sum+=$3} END { print "Average = ",sum/NR}'
SAM/Sequence Alignment Format and BAM format specification
- https://samtools.github.io/hts-specs/SAMv1.pdf and samtools webpage.
- http://genome.sph.umich.edu/wiki/SAM
Germline vs Somatic mutation
Germline: inherit from parents. See the Wikipedia page.
Driver vs passenger mutation
https://en.wikipedia.org/wiki/Somatic_evolution_in_cancer
Simulate Whole genome
- DWGSIM mentioned by Variant Callers for Next-Generation Sequencing Data: A Comparison Study. For its usage, see http://davetang.org/wiki/tiki-index.php?page=DWGSIM
Allele vs Gene
http://www.diffen.com/difference/Allele_vs_Gene
- A gene is a stretch of DNA or RNA that determines a certain trait.
- Genes mutate and can take two or more alternative forms; an allele is one of these forms of a gene. For example, the gene for eye color has several variations (alleles) such as an allele for blue eye color or an allele for brown eyes.
- An allele is found at a fixed spot on a chromosome?
- Chromosomes occur in pairs so organisms have two alleles for each gene — one allele in each chromosome in the pair. Since each chromosome in the pair comes from a different parent, organisms inherit one allele from each parent for each gene. The two alleles inherited from parents may be same (homozygous) or different (heterozygotes).
Locus
https://en.wikipedia.org/wiki/Locus_(genetics)
Haplotypes
- http://www.brown.edu/Research/Istrail_Lab/proj_cmsh.php
- http://www.nature.com/nri/journal/v5/n1/fig_tab/nri1532_F2.html
- https://www.sciencenews.org/article/seeking-genetic-fate
- http://www.medscape.com/viewarticle/553400_3
Base quality, Mapping quality, Variant quality
- Fastq base quality: https://en.wikipedia.org/wiki/FASTQ_format
- Mapping quality: http://genome.cshlp.org/content/18/11/1851.long
- Variant quality: http://www.ncbi.nlm.nih.gov/pubmed/21903627
Nonsynonymous mutation
It is related to the genetic code.
See
- http://evolution.about.com/od/Overview/a/Synonymous-Vs-Nonsynonymous-Mutations.htm
- https://en.wikipedia.org/wiki/Nonsynonymous_substitution
Other software
Partek
- Partek Flow Software http://youtu.be/-6aeQPOYuHY
dCHIP
MeV
MeV v4.8 (11/18/2011) allows annotation from Bioconductor
IPA from Ingenuity
Login: There are web started version https://analysis.ingenuity.com/pa and Java applet version https://analysis.ingenuity.com/pa/login/choice.jsp. We can double click the file <IpaApplication.jnlp> in my machine's download folder.
Features:
- easily search the scientific literature/integrate diverse biological information.
- build dynamic pathway models
- quickly analyze experimental data/Functional discovery: assign function to genes
- share research and collaborate. On the other hand, IPA is web based, so it takes time for running analyses. Once submitted analyses are done, an email will be sent to the user.
Start Here
Expression data -> New core analysis -> Functions/Diseases -> Network analysis
Canonical pathways |
| |
Simple or advanced search --------------------+ |
| |
v |
My pathways, Lists <------+
^
|
Creating a custom pathway --------------------+
Resource:
- http://bioinformatics.mdanderson.org/MicroarrayCourse/Lectures09/Pathway%20Analysis.pdf
- http://libguides.mit.edu/content.php?pid=14149&sid=843471
- http://people.mbi.ohio-state.edu/baguda/PathwayAnalysis/
- IPA 5.5 manual http://people.mbi.ohio-state.edu/baguda/PathwayAnalysis/ipa_help_manual_5.5_v1.pdf
- Help and supports
- Tutorials which includes
- Search for genes
- Analysis results
- Upload and analyze example data
- Upload and analyze your own expression data
- Visualize connections among genes
- Learn more special features
- Human isoform view
- Transcription factor analysis
- Downstream effects analysis
Notes:
- The input data file can be an Excel file with at least one gene ID and expression value at the end of columns (just what BRB-ArrayTools requires in general format importer).
- The data to be uploaded (because IPA is web-based; the projects/analyses will not be saved locally) can be in different forms. See http://ingenuity.force.com/ipa/articles/Feature_Description/Data-Upload-definitions. It uses the term Single/Multiple Observation. An Observation is a list of molecule identifiers and their corresponding expression values for a given experimental treatment. A dataset file may contain a single observation or multiple observations. A Single Observation dataset contains only one experimental condition (i.e. wild-type). A Multiple Observation dataset contains more than one experimental condition (i.e. a time course experiment, a dose response experiment, etc) and can be uploaded into IPA in a single file (e.g. Excel). A maximum of 20 observations in a single file may be uploaded into IPA.
- The instruction http://ingenuity.force.com/ipa/articles/Feature_Description/Data-Upload-definitions shows what kind of gene identifier types IPA accepts.
- In this prostate example data tutorial, the term 'fold change' was used to replace log2 gene expression. The tutorial also uses 1.5 as the fold change expression cutoff.
- The gene table given on the analysis output contains columns 'Fold change', 'ID', 'Notes', 'Symbol' (with tooltip), 'Entrez Gene Name', 'Location', 'Types', 'Drugs'. See a screenshot below.
Screenshots:
DAVID Bioinformatics Resource
It offers an integrated annotation combining gene ontology, pathways and protein annotations.
qpcR
Model fitting, optimal model selection and calculation of various features that are essential in the analysis of quantitative real-time polymerase chain reaction (qPCR).