Anders2013
The data is used in the paper "Count-based differential ....." by Anders et al 2013. The paper has been cited 8136 times according to scholar.google.com and 4330 times according to PubMed on Feb 7, 2019.
This page focuses on RNA-Seq. For DNA-Seq see the Genome page.
Required tools
The version number indicated below is the one I use. It may be updated when you are ready to download that.
- bowtie (bowtie2-2.1.0-linux-x86_64.zip): Mac, Linux, Windows binaries
- samtools (samtools-0.1.19.tar.bz2): require GCC to compile (build-essential) & zlib1g-dev & libncurses5-dev packages. wikipedia.
- tophat (tophat-2.0.10.Linux_x86_64.tar.gz): Linux and Mac binaries. Mailing list is on Tuxedo Tools Users Google Group.
- sra toolkit (sratoolkit.2.3.4-2-ubuntu64.tar.gz): Mac, Ubuntu & CentOS binaries
- HTseq (HTSeq-0.6.1.tar.gz): require Python 2 and pysam package
- IGV (IGV_2.3.26.zip): require Java and registration to download the program
- FastQC (fastqc_v0.10.1.zip): require Java (jdk is needed, jre is not enough)
- FastX (fastx_toolkit_0.0.13_binaries_Linux_2.6_amd64.tar.bz2): require libgtextutils-0.6
Note
- For alignment, STAR is another choice.
- For trimmer, trimmomatic is another choice. For example, we can use it to remove low quality reads (<30).
java -jar trimmomatic-0.36.jar -phred33 -threads 8 file1.fastq.gz file2.fastq.gz -baseout file.fastq.gz AVGQUAL:30 java -jar trimmomatic-0.36.jar -phred33 -threads 8 file1.fastq.gz file2.fastq.gz -baseout file.fastq.gz HEADCROP:5 MINLEN:50 AVGQUAL:20
Installation
If binary executables are available, we don't need to do anything special except adding the path containing the binary to the PATH variable.
If only source code is available, make sure the required tool GCC is installed (Under Ubuntu, we can use sudo apt-get install build-essential). Then we can compile and then install the program by using ./configure, make and make install.
In this exercise, all source code or binary programs were extracted to $HOME/binary/ directory (Use mkdir ~/binary to create this new directory) where each program has its own directory.
To use a shell script to install every thing, try the following.
cd; wget -N https://www.dropbox.com/s/ip1jiarxhq0dq91/install.sh; chmod +x install.sh; sudo ./install.sh; rm install.sh
Change PATH variable
Open ~/.basrhc file using any text editor (such as nano or vi) and add the path containing the binary of each program to PATH variable.
tail ~/.bashrc export PATH=$PATH:~/binary/bowtie2-2.1.0/ export PATH=$PATH:~/binary/tophat-2.0.10.Linux_x86_64/ export PATH=$PATH:~/binary/samtools-0.1.19/ export PATH=$PATH:~/binary/samtools-0.1.19/bcftools/ export PATH=$PATH:~/binary/samtools-0.1.19/misc/ export PATH=$PATH:~/binary/sratoolkit.2.3.4-2-ubuntu64/bin export PATH=$PATH:~/binary/HTSeq-0.6.1/build/scripts-2.7 export PATH=$PATH:~/binary/IGV_2.3.26/ export PATH=$PATH:~/binary/FastQC/ export PATH=$PATH:~/binary/fastx/bin
If we want to make these tools available for ALL users (ie system wide environment), we need to put these lines on /etc/profile file.
HTSeq and pysam
HTSeq requires Python 2 and pysam package. For pysam package, we have two ways to install it
1. Use pip
sudo apt-get install python-pip # sudo -H pip install pysam sudo -H pip install pysam==0.10.0 # lock the version
The "-H" flag is used to suppress a warning message.
The latest version of pysam may fail so it is not good to use the command pip install pysam.
$ python
>>> import pysam
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/Library/Python/2.7/site-packages/pysam/__init__.py", line 5, in <module>
from pysam.libchtslib import *
File "pysam/libchtslib.pyx", line 1, in init pysam.libchtslib (pysam/libchtslib.c:12515)
ImportError: dlopen(/Library/Python/2.7/site-packages/pysam/libcutils.so, 2): Library not loaded: @rpath/pysam-0.11.2.2-py2.7-macosx-10.12-intel.egg/pysam/libcsamtools.so
Referenced from: /Library/Python/2.7/site-packages/pysam/libcutils.so
Reason: image not found
2. Use python to build/install.
wget https://pypi.python.org/packages/de/03/02934438b204565bc5231f38a11da840a3c3e4b2beac8c8770d675770668/pysam-0.9.1.4.tar.gz tar xzvf pysam-0.9.1.4.tar.gz sudo apt-get -y install zlib1g-dev sudo apt-get install python-setuptools python setup.py build sudo python setup.py install # Installed /usr/local/lib/python2.7/dist-packages/pysam-0.9.1.4-py2.7-linux-x86_64.egg
Data directory
Download <nprot.2013.099-S1.zip> from the paper's web site and extract it to ~/Anders2013 directory.
brb@brbweb4:~$ mkdir Anders2013 brb@brbweb4:~$ cd Anders2013 brb@brbweb4:~/Anders2013$ wget http://www.nature.com/nprot/journal/v8/n9/extref/nprot.2013.099-S1.zip brb@brbweb4:~/Anders2013$ unzip nprot.2013.099-S1.zip brb@brbweb4:~/Anders2013$ ls CG8144_RNAi-1.count counts.csv README.txt Untreated-1.count Untreated-6.count CG8144_RNAi-3.count __MACOSX samples.csv Untreated-3.count CG8144_RNAi-4.count nprot.2013.099-S1.zip SraRunInfo.csv Untreated-4.count
The raw data is from GSE18508 'modENCODE Drosophila RNA Binding Protein RNAi RNA-Seq Studies'.
Subdirectory
During running tophat program, several subdirectories will be generated. Each subdirectory will contain accepted_hits.bam, junctions.bed, insertions.bed, deletions.bed and other files/sub-subdirectories.
Once samtools program was run, .bam (and .bai) files will be created under Anders2013 directory.
Pipeline
The following summary is extracted from http://www.bioconductor.org/help/course-materials/2013/CSAMA2013/tuesday/afternoon/DESeq_protocol.pdf. It is the same as Anders 2013 paper but the paper requires an access permission.
It would be interesting to monitor the disk space usage before, during and after the execution.
Other pipelines:
- RNA-Seq Data Analysis: From Raw Data Quality Control to Differential Expression Analysis, Sep 2017. Bash and R scripts are included.
Download example data (22 files in SRA format)
Download Supplement file and read <SraRunInfo.csv> in R. We will choose a subset of data based on "LibraryName" column.
sri = read.csv("SraRunInfo.csv", stringsAsFactors=FALSE)
keep = grep("CG8144|Untreated-",sri$LibraryName)
sri = sri[keep,]
fs = basename(sri$download_path)
for(i in 1:nrow(sri))
download.file(sri$download_path[i], fs[i])
names(sri)
[1] "Run" "ReleaseDate" "spots" "bases"
[5] "avgLength" "size_MB" "AssemblyName" "download_path"
[9] "Experiment" "LibraryName" "LibraryStrategy" "LibrarySelection"
[13] "LibrarySource" "LibraryLayout" "InsertSize" "Platform"
[17] "Model" "SRAStudy" "BioProject" "Study_Pubmed_id"
[21] "ProjectID" "Sample" "BioSample" "TaxID"
[25] "ScientificName" "SampleName" "Submission" "Consent"
apply(sri,2, function(x) length(unique(x)))
Run ReleaseDate spots bases
22 22 22 22
avgLength size_MB AssemblyName download_path
5 22 1 22
Experiment LibraryName LibraryStrategy LibrarySelection
7 7 1 1
LibrarySource LibraryLayout InsertSize Platform
1 2 2 1
Model SRAStudy BioProject Study_Pubmed_id
1 1 1 1
ProjectID Sample BioSample TaxID
1 7 1 1
ScientificName SampleName Submission Consent
1 1 1 1
sri[1:2,]
Run ReleaseDate spots bases avgLength size_MB
1 SRR031714 2010-01-15 10:42:00 5327425 394229450 74 396
2 SRR031715 2010-01-21 14:24:18 5248396 388381304 74 390
AssemblyName
1 NA
2 NA
download_path
1 ftp://ftp-private.ncbi.nlm.nih.gov/sra/sra-instant/reads/ByRun/sra/SRR/SRR031/SRR031714/SRR031714.sra
2 ftp://ftp-private.ncbi.nlm.nih.gov/sra/sra-instant/reads/ByRun/sra/SRR/SRR031/SRR031715/SRR031715.sra
Experiment LibraryName LibraryStrategy LibrarySelection LibrarySource
1 SRX014459 Untreated-3 RNA-Seq cDNA TRANSCRIPTOMIC
2 SRX014459 Untreated-3 RNA-Seq cDNA TRANSCRIPTOMIC
LibraryLayout InsertSize Platform Model SRAStudy
1 PAIRED 200 ILLUMINA Illumina Genome Analyzer II SRP001537
2 PAIRED 200 ILLUMINA Illumina Genome Analyzer II SRP001537
BioProject Study_Pubmed_id ProjectID Sample BioSample
1 GEO Series accession: GSE18508 20921232 168994 SRS008447 NA
2 GEO Series accession: GSE18508 20921232 168994 SRS008447 NA
TaxID ScientificName SampleName Submission Consent
1 7227 Drosophila melanogaster Drosophila melanogaster SRA010243 public
2 7227 Drosophila melanogaster Drosophila melanogaster SRA010243 public
Convert SRA to FASTQ (sra -> fastq)
(Update) fasterq-dump command. Note there is no --gzip option as in fastq-dump. Use gzip to do that.
This takes a long time and it is not to run in parallel by default. At the end of execution, there are 30 fastq files because 8 SRA file are pair end.
stopifnot( all(file.exists(fs)) ) # assure FTP download was successful
for(f in fs) {
cmd = paste("fastq-dump --split-3", f)
cat(cmd,"\n")
system(cmd) # invoke command
}
fastq-dump --split-3 SRR031714.sra
Read 5327425 spots for SRR031714.sra
Written 5327425 spots for SRR031714.sra
fastq-dump --split-3 SRR031715.sra
Read 5248396 spots for SRR031715.sra
Written 5248396 spots for SRR031715.sra
...
Note that '--split-3' option will splits mate-pair reads into separate files (First biological reads satisfying dumping conditions are placed in files *_1.fastq and *_2.fastq If only one biological read is present it is placed in *.fastq Biological reads and above are ignored). See this post.
brb@brbweb4:~/Anders2013$ ls -lh *.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:47 SRR031708.fastq -rw-r--r-- 1 brb brb 812M Feb 10 12:50 SRR031709.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:56 SRR031710.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 13:02 SRR031711.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:08 SRR031712.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:14 SRR031713.fastq -rw-r--r-- 1 brb brb 1.1G Feb 10 11:48 SRR031714_1.fastq -rw-r--r-- 1 brb brb 1.1G Feb 10 11:48 SRR031714_2.fastq -rw-r--r-- 1 brb brb 1.1G Feb 10 11:54 SRR031715_1.fastq -rw-r--r-- 1 brb brb 1.1G Feb 10 11:54 SRR031715_2.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:02 SRR031716_1.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:02 SRR031716_2.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:10 SRR031717_1.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:10 SRR031717_2.fastq -rw-r--r-- 1 brb brb 1.4G Feb 10 13:21 SRR031718.fastq -rw-r--r-- 1 brb brb 927M Feb 10 13:25 SRR031719.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:32 SRR031720.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:39 SRR031721.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 13:45 SRR031722.fastq -rw-r--r-- 1 brb brb 842M Feb 10 13:49 SRR031723.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:17 SRR031724_1.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:17 SRR031724_2.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:25 SRR031725_1.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:25 SRR031725_2.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:33 SRR031726_1.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 12:33 SRR031726_2.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:41 SRR031727_1.fastq -rw-r--r-- 1 brb brb 1.2G Feb 10 12:41 SRR031727_2.fastq -rw-r--r-- 1 brb brb 1.3G Feb 10 13:54 SRR031728.fastq -rw-r--r-- 1 brb brb 3.2G Feb 10 14:07 SRR031729.fastq
FASTQ PAIR
FASTQ PAIR Rewrite paired end fastq files to make sure that all reads have a mate and to separate out singletons
Access quality control through ShortRead or FastQC
library("ShortRead")
fqQC = qa(dirPath=".", pattern=".fastq$", type="fastq")
report(fqQC, type="html", dest="fastqQAreport")
Use a web browser to inspect the generated HTML file (here, stored in the \fastqQAreport" directory) with the quality-assessment report.
FastQC can be run either interactively or non-interactive. For example
/opt/RNA-Seq/bin/FastQC/fastqc ~/GSE11209/SRR002058.fastq --outdir=/home/brb/GSE11209/fastQC
will create a zip file and an unzipped directory under /home/brb/GSE11209/fastQC.
FastX_trimmer is a better choice. The syntax is
fastx_trimmer -f N -l N -i INPUT -o OUTPUT -v -Q33
The last parameter -Q33 is necessary for Illumina encoding quality score. If we omit it, we will get an error fastx_trimmer: Invalid quality score value (char '#' ord 35 quality value -29) on line 4
Read quality issues
http://training.bioinformatics.ucdavis.edu/docs/2012/05/RNA/qa-and-i.html
- Per base sequence quality (by position)
- Per sequence quality scores: see if a subset of sequences have universally low quality values.
- Per base sequence content
- Per sequence GC content
- Per base N content
- Sequence Length Distribution
- Sequence Duplication Levels
- Overpresented sequences
- Kmer Content
Encoding
- Encoding Starting in Illumina 1.8, the quality scores have basically returned to the use of the Sanger format (Phred+33).
- all the FASTQ sequences from ncbi are Phred+33 (same as Sanger).
Download reference genome (for Tophat)
wget ftp://ftp.ensembl.org/pub/release-70/fasta/drosophila_melanogaster/dna/Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa.gz gunzip Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa.gz
Note that bowtie2/tophat2 indices for many commonly used reference genomes can be downloaded directly from https://ccb.jhu.edu/software/tophat/igenomes.shtml. Actually the link https://support.illumina.com/sequencing/sequencing_software/igenome.html from illumina is more complete.
For human species, it has different assemblies
- Ensembl - GRCh37 (equivalent to hg19)
- NCBI - GRCh38, build37.1, build37.2, build36.3 (equivalent to hg18)
- UCSC - hg38, hg19, hg18
| Source | |||
|---|---|---|---|
| Ensembl | GRCh37 | ||
| NCBI | GRCh38 | build37 | build36 |
| UCSC | hg38 | hg19 | hg18 |
Structure of downloaded reference genome from Tophat
Downloaded genome from tophat takes a long time (20GB for human). Using bowtie2-build program which can takes an enormous time to run.
$ tar xzvf Homo_sapiens_Ensembl_GRCh37.tar.gz
$ tree -L 4 Homo_sapiens
Homo_sapiens
└── Ensembl
└── GRCh37
├── Annotation
│ ├── Archives <==== gtf file
│ ├── Genes -> Archives/archive-current/Genes
│ ├── README.txt -> Archives/archive-current/README.txt
│ ├── SmallRNA -> Archives/archive-current/SmallRNA
│ └── Variation -> Archives/archive-current/Variation
├── GenomeStudio
│ ├── Archives
│ ├── Homo_sapiens -> Archives/archive-2012-03-09-04-49-46/Homo_sapiens
│ └── README.txt
└── Sequence
├── AbundantSequences
├── Bowtie2Index <==== bt2 files
├── BowtieIndex
├── BWAIndex
├── Chromosomes
├── Squashed-Homo_sapiens-Ensembl-GRCh37
└── WholeGenomeFasta <==== fa file
18 directories, 2 files
$ ls -l Homo_sapiens/Ensembl/GRCh37/Annotation/Archives/archive-2013-03-06-14-23-04/Genes
total 639136
-rwxrwxr-x 1 brb brb 302 Mar 18 2013 ChromInfo.txt
-rwxrwxr-x 1 brb brb 603096976 Mar 18 2013 genes.gtf
-rwxrwxr-x 1 brb brb 8236175 Mar 18 2013 refFlat.txt.gz
-rwxrwxr-x 1 brb brb 43130092 Mar 18 2013 refGene.txt
A list of files in <Homo_sapiens_UCSC_hg18.tar.gz> can be found on (File:Ucsc hg18.txt) or <Homo_sapiens_UCSC_hg19.tar.gz> can be found on github.
We don't need to extract the whole tarball. We just need to extract some files we need. But how do we know the locations of files we need? Use tar -tzvf XXX.tar.gz to find out.
$ tar -xzvf Homo_sapiens_UCSC_hg18.tar.gz Homo_sapiens/UCSC/hg18/Annotation/Archives/archive-2014-06-02-13-47-56/Genes/genes.gtf
Homo_sapiens/UCSC/hg18/Annotation/Archives/archive-2014-06-02-13-47-56/Genes/genes.gtf
$ tar -xzvf Homo_sapiens_UCSC_hg18.tar.gz Homo_sapiens/UCSC/hg18/Sequence/Bowtie2Index/
Homo_sapiens/UCSC/hg18/Sequence/Bowtie2Index/
Homo_sapiens/UCSC/hg18/Sequence/Bowtie2Index/genome.3.bt2
Homo_sapiens/UCSC/hg18/Sequence/Bowtie2Index/genome.1.bt2
Homo_sapiens/UCSC/hg18/Sequence/Bowtie2Index/genome.rev.2.bt2
Homo_sapiens/UCSC/hg18/Sequence/Bowtie2Index/genome.rev.1.bt2
Homo_sapiens/UCSC/hg18/Sequence/Bowtie2Index/genome.4.bt2
Homo_sapiens/UCSC/hg18/Sequence/Bowtie2Index/genome.2.bt2
Homo_sapiens/UCSC/hg18/Sequence/Bowtie2Index/genome.fa
$ tar -xzvf Homo_sapiens_UCSC_hg18.tar.gz Homo_sapiens/UCSC/hg18/Sequence/WholeGenomeFasta/genome.fa
Homo_sapiens/UCSC/hg18/Sequence/WholeGenomeFasta/genome.fa
$ tree -L 7 Homo_sapiens
Homo_sapiens
└── hg18
├── Annotation
│ └── Archives
│ └── archive-2014-06-02-13-47-56
│ └── Genes
│ └── genes.gtf <= gtf file
└── Sequence
├── Bowtie2Index
│ ├── genome.1.bt2 <= bt2 file
│ ├── genome.2.bt2
│ ├── genome.3.bt2
│ ├── genome.4.bt2
│ ├── genome.fa -> ../WholeGenomeFasta/genome.fa <= fake fa file (symbolic link)
│ ├── genome.rev.1.bt2
│ └── genome.rev.2.bt2
└── WholeGenomeFasta
└── genome.fa <= fa file
Download GTF (gene transfer format) file (for Tophat)
The compressed file is 77.6 MB size.
wget ftp://ftp.ensembl.org/pub/release-70/gtf/drosophila_melanogaster/Drosophila_melanogaster.BDGP5.70.gtf.gz gunzip Drosophila_melanogaster.BDGP5.70.gtf.gz
CRITICAL: Make sure that the gene annotation uses the same coordinate system as the reference FASTA le. Here, both les use BDGP5 (i. e., release 5 of the assembly provided by the Berkeley Drosophila Genome Project), as is apparent from the le names. To be on the safe side here, we recommend to always download the FASTA reference sequence and the GTF annotation data from the same resource provider.
- Ensembl.org provides both fasta and gtf files together. It's useful when we want to use Galaxy.
- Broad also provides a link to download gtf and fasta file.
Preprocess reference FASTA files into an index (fasta -> bt2)
Alternatively, pre-built indices for many commonly-used genomes are available from http://tophat.cbcb.umd.edu/igenomes.html <Drosophila_melanogaster_Ensembl_BDGP5.tar.gz> contains genome.1.bt2, genome.2.bt2, genome.3.bt2, genome.fa, genome.rev.1.bt2, and genome.rev.2.bt2 under Drosophila_melanogaster/Ensembl/BDGP5/Sequence/Bowtie2Index/ directory. This takes about 4 minutes 37 seconds on my Core-i7 Laptop computer running xubuntu 12.04.
When I try to run bowtie2-build on mouse genome, it is extremely slow. Unfortunately bowtie2-build does not have "-p" option like bowtie2 to speed it up.
bowtie2-build -f Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa Dme1_BDGP5_70 brb@brbweb4:~/Anders2013$ ls -lt *.bt2 -rw-r--r-- 1 brb brb 58770281 Feb 10 12:06 Dme1_BDGP5_70.rev.1.bt2 -rw-r--r-- 1 brb brb 40591960 Feb 10 12:06 Dme1_BDGP5_70.rev.2.bt2 -rw-r--r-- 1 brb brb 58770281 Feb 10 12:04 Dme1_BDGP5_70.1.bt2 -rw-r--r-- 1 brb brb 40591960 Feb 10 12:04 Dme1_BDGP5_70.2.bt2 -rw-r--r-- 1 brb brb 339200 Feb 10 12:01 Dme1_BDGP5_70.3.bt2 -rw-r--r-- 1 brb brb 40591953 Feb 10 12:01 Dme1_BDGP5_70.4.bt2
Create a metadata table called "samples"
sri$LibraryName = gsub("S2_DRSC_","",sri$LibraryName) # trim label
samples = unique(sri[,c("LibraryName","LibraryLayout")])
for(i in seq_len(nrow(samples))) {
rw = (sri$LibraryName==samples$LibraryName[i])
if(samples$LibraryLayout[i]=="PAIRED") {
samples$fastq1[i] = paste0(sri$Run[rw],"_1.fastq",collapse=",")
samples$fastq2[i] = paste0(sri$Run[rw],"_2.fastq",collapse=",")
} else {
samples$fastq1[i] = paste0(sri$Run[rw],".fastq",collapse=",")
samples$fastq2[i] = ""
}
}
samples$condition = "CTL"
samples$condition[grep("RNAi",samples$LibraryName)] = "KD"
samples$shortname = paste( substr(samples$condition,1,2),
substr(samples$LibraryLayout,1,2), seq_len(nrow(samples)), sep=".")
samples
LibraryName LibraryLayout
1 Untreated-3 PAIRED
3 Untreated-4 PAIRED
5 CG8144_RNAi-3 PAIRED
7 CG8144_RNAi-4 PAIRED
144 Untreated-1 SINGLE
150 CG8144_RNAi-1 SINGLE
156 Untreated-6 SINGLE
fastq1
1 SRR031714_1.fastq,SRR031715_1.fastq
3 SRR031716_1.fastq,SRR031717_1.fastq
5 SRR031724_1.fastq,SRR031725_1.fastq
7 SRR031726_1.fastq,SRR031727_1.fastq
144 SRR031708.fastq,SRR031709.fastq,SRR031710.fastq,SRR031711.fastq,SRR031712.fastq,SRR031713.fastq
150 SRR031718.fastq,SRR031719.fastq,SRR031720.fastq,SRR031721.fastq,SRR031722.fastq,SRR031723.fastq
156 SRR031728.fastq,SRR031729.fastq
fastq2 condition shortname
1 SRR031714_2.fastq,SRR031715_2.fastq CTL CT.PA.1
3 SRR031716_2.fastq,SRR031717_2.fastq CTL CT.PA.2
5 SRR031724_2.fastq,SRR031725_2.fastq KD KD.PA.3
7 SRR031726_2.fastq,SRR031727_2.fastq KD KD.PA.4
144 CTL CT.SI.5
150 KD KD.SI.6
156 CTL CT.SI.7
On the other hand, the <samples.txt> file used in BRB-DGE looks like
$ cat samples.txt LibraryName LibraryLayout fastq1 fastq2 Untreated-3 PAIRED SRR031714_1.fastq,SRR031715_1.fastq SRR031714_2.fastq,SRR031715_2.fastq Untreated-4 PAIRED SRR031716_1.fastq,SRR031717_1.fastq SRR031716_2.fastq,SRR031717_2.fastq CG8144_RNAi-3 PAIRED SRR031724_1.fastq,SRR031725_1.fastq SRR031724_2.fastq,SRR031725_2.fastq CG8144_RNAi-4 PAIRED SRR031726_1.fastq,SRR031727_1.fastq SRR031726_2.fastq,SRR031727_2.fastq Untreated-1 SINGLE SRR031708.fastq,SRR031709.fastq,SRR031710.fastq,SRR031711.fastq,SRR031712.fastq,SRR031713.fastq CG8144_RNAi-1 SINGLE SRR031718.fastq,SRR031719.fastq,SRR031720.fastq,SRR031721.fastq,SRR031722.fastq,SRR031723.fastq Untreated-6 SINGLE SRR031728.fastq,SRR031729.fastq
Align the reads to the reference genome using tophat2 (fastq -> bam, bed)
Note:
- '-p' specifies the number of threads to use, -o specifies the output directory. The first name (bowind) is the name of the index (built in advance).
- It seems even the program cannot find the reference genome, it still runs.
- The run time is about 3 to 4 hours per sample when 7 jobs were running at the same time (5 threads for each sample).
- If only one sample was run it took 45 minutes as the paper said (5 threads). If I specify 1 thread, the running time is 1 hour 44 minutes.
- If I only run 2 jobs using batch, it takes 1 hour and 1 hour 2 mintues to finish these two jobs.
- The input is fastq files. The output is a directory contains 7 files (including *.bam) and 1 subdirectory of logs.
gf = "Drosophila_melanogaster.BDGP5.70.gtf"
bowind = "Dme1_BDGP5_70" # bt2 files
cmd = with(samples, paste("tophat2 -G", gf, "-p 5 -o", LibraryName,
bowind, fastq1, fastq2))
cmd # Actually we want to use system() to run each line of cmd.
[1] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o Untreated-3 Dme1_BDGP5_70 SRR031714_1.fastq,SRR031715_1.fastq SRR031714_2.fastq,SRR031715_2.fastq"
[2] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o Untreated-4 Dme1_BDGP5_70 SRR031716_1.fastq,SRR031717_1.fastq SRR031716_2.fastq,SRR031717_2.fastq"
[3] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o CG8144_RNAi-3 Dme1_BDGP5_70 SRR031724_1.fastq,SRR031725_1.fastq SRR031724_2.fastq,SRR031725_2.fastq"
[4] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o CG8144_RNAi-4 Dme1_BDGP5_70 SRR031726_1.fastq,SRR031727_1.fastq SRR031726_2.fastq,SRR031727_2.fastq"
[5] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o Untreated-1 Dme1_BDGP5_70 SRR031708.fastq,SRR031709.fastq,SRR031710.fastq,SRR031711.fastq,SRR031712.fastq,SRR031713.fastq "
[6] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o CG8144_RNAi-1 Dme1_BDGP5_70 SRR031718.fastq,SRR031719.fastq,SRR031720.fastq,SRR031721.fastq,SRR031722.fastq,SRR031723.fastq "
[7] "tophat2 -G Drosophila_melanogaster.BDGP5.70.gtf -p 5 -o Untreated-6 Dme1_BDGP5_70 SRR031728.fastq,SRR031729.fastq "
Tophat options
- --coverage-search/--no-coverage-search: According to this, it is a step to define possible junctions between exons. If you only want the expression profile of the annotated genes, you can skip this step for speed. The identification of new regions with the coverage is relevant only if you want to detect new splice sites in alternate transcripts or even new genes.
- -r/--mate-inner-dist <int>: This is the expected (mean) inner distance between mate pairs. For, example, for paired end runs with fragments selected at 300bp, where each end is 50bp, you should set -r to be 200. The default is 50bp. Check out this post about fragment size & inner size. This post discusses about the fragment size.
- -g/--max-multihits <int>: Instructs TopHat to allow up to this many alignments to the reference for a given read, and choose the alignments based on their alignment scores if there are more than this number. The default is 20 for read mapping.
- --library-type: The default is unstranded (fr-unstranded). If either fr-firststrand or fr-secondstrand is specified, every read alignment will have an XS attribute tag.
- --GTF option: Tophat now takes the approach of mapping the reads on the transcriptome first, with only the unmapped reads being further aligned to the whole genome and going through the novel junction discovery process like before. Please note that the values in the first column of the provided GTF/GFF file (column which indicates the chromosome or contig on which the feature is located), must match the name of the reference sequence in the Bowtie index you are using with TopHat.
- -T/--transcriptome-only: only align the reads to the transcriptome and report only those mappings as genomic mappings. Note that we still get junctions map.
- -x/--transcriptome-max-hits: Maximum number of mappings allowed for a read, when aligned to the transcriptome (any reads found with more then this number of mappings will be discarded).
Some helpful information:
Output files
$ ls -lh Untreated-6 total 2.2G -rw-rw-r-- 1 brb brb 2.0G May 6 18:11 accepted_hits.bam -rw-rw-r-- 1 brb brb 202 May 6 18:03 align_summary.txt -rw-rw-r-- 1 brb brb 2.0M May 6 18:03 deletions.bed -rw-rw-r-- 1 brb brb 612K May 6 18:03 insertions.bed -rw-rw-r-- 1 brb brb 2.7M May 6 18:03 junctions.bed drwxrwxr-x 2 brb brb 4.0K May 6 18:11 logs -rw-rw-r-- 1 brb brb 70 May 6 17:43 prep_reads.info -rw-rw-r-- 1 brb brb 197M May 6 18:12 unmapped.bam $ ls -lh Untreated-6/logs total 108K -rw-rw-r-- 1 brb brb 0 May 6 18:11 bam_merge_um.log -rw-rw-r-- 1 brb brb 12K May 6 17:59 bowtie_build.log -rw-rw-r-- 1 brb brb 0 May 6 17:39 bowtie_inspect_recons.log -rw-rw-r-- 1 brb brb 221 May 6 17:53 bowtie.left_kept_reads.log -rw-rw-r-- 1 brb brb 216 May 6 17:59 bowtie.left_kept_reads_seg1.log -rw-rw-r-- 1 brb brb 216 May 6 17:59 bowtie.left_kept_reads_seg2.log -rw-rw-r-- 1 brb brb 215 May 6 18:00 bowtie.left_kept_reads_seg3.log -rw-rw-r-- 1 brb brb 394 May 6 17:59 juncs_db.log -rw-rw-r-- 1 brb brb 300 May 6 18:00 long_spanning_reads.segs.log -rw-rw-r-- 1 brb brb 105 May 6 17:43 prep_reads.log -rw-rw-r-- 1 brb brb 617 May 6 18:03 reports.log -rw-rw-r-- 1 brb brb 0 May 6 18:07 reports.merge_bam.log -rw-rw-r-- 1 brb brb 40 May 6 18:05 reports.samtools_sort.log0 -rw-rw-r-- 1 brb brb 40 May 6 18:05 reports.samtools_sort.log1 -rw-rw-r-- 1 brb brb 40 May 6 18:04 reports.samtools_sort.log10 -rw-rw-r-- 1 brb brb 40 May 6 18:05 reports.samtools_sort.log2 -rw-rw-r-- 1 brb brb 40 May 6 18:04 reports.samtools_sort.log3 -rw-rw-r-- 1 brb brb 40 May 6 18:06 reports.samtools_sort.log4 -rw-rw-r-- 1 brb brb 40 May 6 18:06 reports.samtools_sort.log5 -rw-rw-r-- 1 brb brb 40 May 6 18:05 reports.samtools_sort.log6 -rw-rw-r-- 1 brb brb 40 May 6 18:05 reports.samtools_sort.log7 -rw-rw-r-- 1 brb brb 40 May 6 18:04 reports.samtools_sort.log8 -rw-rw-r-- 1 brb brb 40 May 6 18:04 reports.samtools_sort.log9 -rw-rw-r-- 1 brb brb 12K May 6 18:12 run.log -rw-rw-r-- 1 brb brb 813 May 6 17:58 segment_juncs.log -rw-rw-r-- 1 brb brb 2.0K May 6 18:12 tophat.log
transcriptome_data
If we specify --transcriptome-index=transcriptome_data/known in tophat2 command, we shall get a directory transcriptome with several files
known.1.bt2 known.2.bt2 known.3.bt2 known.4.bt2 known.ver known.fa known.fa.tlst known.gff
Output from console
[2014-04-18 16:42:12] Beginning TopHat run (v2.0.10) ----------------------------------------------- [2014-04-18 16:42:12] Checking for Bowtie Bowtie version: 2.1.0.0 [2014-04-18 16:42:12] Checking for Samtools Samtools version: 0.1.19.0 [2014-04-18 16:42:12] Checking for Bowtie index files (genome).. [2014-04-18 16:42:12] Checking for reference FASTA file [2014-04-18 16:42:12] Generating SAM header for genome [2014-04-18 16:42:58] Reading known junctions from GTF file [2014-04-18 16:43:17] Preparing reads left reads: min. length=49, max. length=49, 26721504 kept reads (2587 discarded) [2014-04-18 16:47:40] Building transcriptome data files E2_Rep2/tmp/genes [2014-04-18 16:48:22] Building Bowtie index from genes.fa [2014-04-18 17:12:33] Mapping left_kept_reads to transcriptome genes with Bowtie2 [2014-04-18 17:44:09] Resuming TopHat pipeline with unmapped reads [2014-04-18 17:44:09] Mapping left_kept_reads.m2g_um to genome genome with Bowtie2 [2014-04-18 17:49:50] Mapping left_kept_reads.m2g_um_seg1 to genome genome with Bowtie2 (1/2) [2014-04-18 17:51:07] Mapping left_kept_reads.m2g_um_seg2 to genome genome with Bowtie2 (2/2) [2014-04-18 17:51:54] Searching for junctions via segment mapping Coverage-search algorithm is turned on, making this step very slow Please try running TopHat again with the option (--no-coverage-search) if this step takes too much time or memory. [2014-04-18 18:08:13] Retrieving sequences for splices [2014-04-18 18:10:10] Indexing splices [2014-04-18 18:19:15] Mapping left_kept_reads.m2g_um_seg1 to genome segment_juncs with Bowtie2 (1/2) [2014-04-18 18:21:26] Mapping left_kept_reads.m2g_um_seg2 to genome segment_juncs with Bowtie2 (2/2) [2014-04-18 18:23:38] Joining segment hits [2014-04-18 18:25:52] Reporting output tracks ----------------------------------------------- [2014-04-18 18:38:05] A summary of the alignment counts can be found in E2_Rep2/align_summary.txt [2014-04-18 18:38:05] Run complete: 01:55:52 elapsed
Some lessons
The following table summarizes the result of using '-G' and '--transcriptome-index' options. In conclusion, 3 possible ways of using these 2 options:
- Not using '-G'
- Using '-G' but no '--transcriptome-index'
- Run tophat2 one time to generate transcriptome index files and then run tophat2 again with '-G' and '--transcriptome-index' options.
| Example 1 (fail) | Wrong spec --transcriptome-index |
| Example 2 (fail) | Using -G & --transcriptome-index but my files (genes.gtf & genome.*) |
| Example 3 (OK) | No -G option |
| Example 4 (OK) | Proper use of using -G & --transcriptome-index (2 steps) |
| Example 5 (fail) | Using -G & --transcriptome-index using Illumina iGenomes dir |
| Example 6 (fail) | Like Example 5 but cp genes.gtf to genome.gtf |
| Example 7 (OK) | Using -G but no --transcriptome-index |
More details can be found on my github website.
To create the transcriptome index folder, it is not necessary to allocate too much memory or multithreads. The 'swarm' and 'jobload' commands show 0.3 GB memory and 1 thread is used even we allocate more memory and more CPUs. It took about 15 minutes to create the folder when the following code is used
tophat2 -p 2 \
-G "/fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf" \
--transcriptome-index=transcriptome_data/known \
"/fdb/igenomes/Homo_sapiens/UCSC/hg19/Sequence/Bowtie2Index/genome"
This post discussed Tophat with and without GTF.
FAQs for Tophat
- TopHat2 on multiple samples, avoid building Bowtie index from genes.fa each time?
Is it necessary to run the following code by Tophat? See this post on seqanswers.com.
[2014-09-18 10:38:45] Building transcriptome data files /tmp/genes [2014-09-18 10:39:21] Building Bowtie index from genes.fa
For other users, which may encounter the same challenge - The trick is to run this command first:
tophat2 -G iGenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf \
--transcriptome-index=transcriptome_data/known \
iGenomes/Homo_sapiens/UCSC/hg19/Sequence/Bowtie2Index/genome
and then subsequently call tophat2 with this command:
tophat2 --num-threads 12 --transcriptome-index=transcriptome_data/known \
iGenomes/Homo_sapiens/UCSC/hg19/Sequence/Bowtie2Index/genome
myfastq_R1.fastq.gz myfastq_R2.fastq.gz
After running the above command, you'll see the following 1 line (instead of 2 lines as shown above)
[2014-09-18 12:12:04] Using pre-built transcriptome data..
Which is significantly faster, when running multiple samples.
The UCSC/hg19 data can retrieved like so:
wget ftp://igenome:[email protected]/Homo_sapiens/UCSC/hg19/Homo_sapiens_UCSC_hg19.tar.gz
Organize, sort and index the BAM files and create SAM files (bam -> sam)
Note
- Potentially we can run the commands in parallel. This step needs lots of disk I/O so it may be better not to run the command in parallel.
- Four steps:
- first step - Input: original aligned bam file. Output: <Untreated-3_sn.bam> (1.5 GB). sort alignment file by read name (-n option).
- Second step - Input: sorted bam file from 1st step. Output: <Untreated-3_sn.sam> (11 GB). BAM<->SAM conversion. The SAM file generated from these 2 steps will be used in HTSEQ-count (For paired-end data, the alignment have to be sorted either by read name or by alignment position; see htseq-count documentation).
- Third step - Input: original aligned bam file. Output: <Untreated-3_s.bam> (1.2 GB). sort alignment file by chromosomal coordinates.
- Fourth step - Input: sorted bam file from 3rd step. Output: <Untreated-3_s.bam.bai> (375 KB). index alignment. The SAM and BAI files generated from these 2 steps together with GTF file will be used in IGV (IGV requires that both SAM and BAM files be sorted by position and indexed; see IGV documentation).
- Examples how to use. See here
If we use samtools view to see the sorted bam files, we may find some fagments (first column) has only one read in this file and some fragments are not in this file.
If we look into the (sorted by name) unmapped.bam file, we can see the unmapped fragments.
for(i in seq_len(nrow(samples))) {
lib = samples$LibraryName[i]
ob = file.path(lib, "accepted_hits.bam")
# sort by name, convert to SAM for htseq-count
cat(paste0("samtools sort -n ",ob," ",lib,"_sn"),"\n")
cat(paste0("samtools view -o ",lib,"_sn.sam ",lib,"_sn.bam"),"\n")
# sort by position and index for IGV
cat(paste0("samtools sort ",ob," ",lib,"_s"),"\n")
cat(paste0("samtools index ",lib,"_s.bam"),"\n\n")
}
samtools sort -n Untreated-3/accepted_hits.bam Untreated-3_sn
samtools view -o Untreated-3_sn.sam Untreated-3_sn.bam
samtools sort Untreated-3/accepted_hits.bam Untreated-3_s
samtools index Untreated-3_s.bam
samtools sort -n Untreated-4/accepted_hits.bam Untreated-4_sn
samtools view -o Untreated-4_sn.sam Untreated-4_sn.bam
samtools sort Untreated-4/accepted_hits.bam Untreated-4_s
samtools index Untreated-4_s.bam
samtools sort -n CG8144_RNAi-3/accepted_hits.bam CG8144_RNAi-3_sn
samtools view -o CG8144_RNAi-3_sn.sam CG8144_RNAi-3_sn.bam
samtools sort CG8144_RNAi-3/accepted_hits.bam CG8144_RNAi-3_s
samtools index CG8144_RNAi-3_s.bam
samtools sort -n CG8144_RNAi-4/accepted_hits.bam CG8144_RNAi-4_sn
samtools view -o CG8144_RNAi-4_sn.sam CG8144_RNAi-4_sn.bam
samtools sort CG8144_RNAi-4/accepted_hits.bam CG8144_RNAi-4_s
samtools index CG8144_RNAi-4_s.bam
samtools sort -n Untreated-1/accepted_hits.bam Untreated-1_sn
samtools view -o Untreated-1_sn.sam Untreated-1_sn.bam
samtools sort Untreated-1/accepted_hits.bam Untreated-1_s
samtools index Untreated-1_s.bam
samtools sort -n CG8144_RNAi-1/accepted_hits.bam CG8144_RNAi-1_sn
samtools view -o CG8144_RNAi-1_sn.sam CG8144_RNAi-1_sn.bam
samtools sort CG8144_RNAi-1/accepted_hits.bam CG8144_RNAi-1_s
samtools index CG8144_RNAi-1_s.bam
samtools sort -n Untreated-6/accepted_hits.bam Untreated-6_sn
samtools view -o Untreated-6_sn.sam Untreated-6_sn.bam
samtools sort Untreated-6/accepted_hits.bam Untreated-6_s
samtools index Untreated-6_s.bam
Output from console
[bam_sort_core] merging from 15 files...
Check Coverage from bam file
Use samtools mpileup or Qualimap software as commented inbiostars.
The output of samtools mpileup has a pileup format. For example, in the GSE37918 dataset,
$ wc -l MDA-MB-231-control_mpileup.txt $ head -n 2 MDA-MB-231-control_mpileup.txt 1 12060 N 1 ^!C B 1 12061 N 1 T C
where each line consists of chromosome, 1-based coordinate, reference base, the number of reads covering the site, read bases and base qualities.
Or use igvtools https://hpc.nih.gov/apps/IGV.html
Inspect alignments with IGV
The Integrative Genomics Viewer (IGV) is a high-performance visualization tool for interactive exploration of large, integrated genomic datasets. It supports a wide variety of data types, including array-based and next-generation sequence data, and genomic annotations.
Launch
I can launch IGV in a terminal (suppose the file IGV_2.3.52.zip is extracted to the directory called binary under the HOME directory):
~/binary/IGV_2.3.52/igv.sh
Note that in addition to the bam file, we need to run samtools index XXX.bam to create the bai file before IGV can use it.
Command line options
bash igv.sh input.bam -g hg19 -b igv_batch_script.txt
And a sample <igv_batch_script.txt> file
new genome hg19 load input.bam snapshotDirectory /path/to/igv_screenshots goto chr19:59418052-59418053 sort base collapse snapshot chr19_59418052-59418053.png goto chr1:15680529-15680532 chr3:5680521-5680533 sort base collapse snapshot chr1_15680529-15680532.chr3_5680521-5680533.png exit
Genomes
IGV contains a lot of genomes. We can access them by using the drop-down list box and select 'More ...' entry. See Hosted Genomes for more information about the meaning of this list (some of them on the list do not appear on the website though).
To create .genome file, following the steps:
- Download reference genome fasta file (bt2 won't work)
wget ftp://ftp.ensembl.org/pub/release-70/fasta/drosophila_melanogaster/dna/Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa.gz gunzip Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa.gz
- Make sure gene model annotation file is available (*.gtf)
- If you have the cytoband file, IGV can display the chromosome ideogram. Otherwise, we won't see the ideogram. Unfortunately I don't see a download choice for the cytoband on the ensembl.org website. The UCSC website does provide the cytoband.txt file for downloading.
- In IGV, click Genomes > Create .genom File. Enter 'Anders2013' for both Unique identifer & Descriptive name. Browse the right FASTA file and Gene file. Click OK. We can save the file <Anders2013.genome> under the data directory.
If the above steps work, we should be able to prepare a sorted bam file with its index file to be used in IGV. For example,
samtools sort Untreated-6/accepted_hits.bam Untreated-6_s samtools index Untreated-6_s.bam
Splice junctions
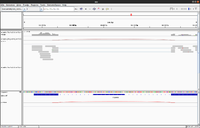
The junctions track calls a splicing event when at least a single read splits across two exons in the alignment track.
IGV defines exons by your sample's read alignments.
Each splice junction is represented by an arc from the beginning to the end of the junction.
- Junctions from the + strand are colored red and extend above the center line.
- Junctions from the – strand are blue and extend below the center line.
See an example of it in IGV from GTF.
The bed file format is specified in genome.ucsc.edu and a few lines of <junctions.bed> are given below.
brb@brb-T3500:~/Anders2013/Untreated-6$ head junctions.bed track name=junctions description="TopHat junctions" 2L 11095 11479 JUNC00000001 4 - 11095 11479 255,0,0 2 74,70 0,314 2L 11271 11479 JUNC00000002 24 - 11271 11479 255,0,0 2 73,70 0,138 2L 11447 11842 JUNC00000003 45 - 11447 11842 255,0,0 2 71,64 0,331
By default IGV won't show the splice junction. Before loading data, check Show junction track in the Alignment Preferences panel. The panel's settings must be adjusted for your data as default settings are for genomic reads for which splicing is irrelevant.
Below is an example from BRB-SeqTools GSE11209 dataset (Saccharomyces_cerevisiae 酵母屬 yeast).
brb@T3600 ~/Downloads/testdata/GSE11209/dT_ori $ cat junctions.bed track name=junctions description="TopHat junctions" I 142233 142632 JUNC00000001 3 + 142233 142632 255,0,0 2 20,13 0,386 II 60161 60721 JUNC00000002 6 - 60161 60721 255,0,0 2 32,24 0,536 II 332859 333403 JUNC00000003 1 + 332859 333403 255,0,0 2 16,17 0,527 ...
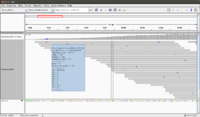
Small test data from Tophat (paired end)
The following screenshot was taken from another (small) dataset with two fq files (paired end). There are 100 sequences in the fq file. The reference genome has only 13 sequences. See also this post.
Before using IGV, we need to run Tophat and samtools (one is to get sorted by position in the reference in bam format, and the other is to get an index file in bai format). Both bam and bai files are binary. If we just use the bam file generated from Tophat in IGV, IGV will show a message 'An index file is required for SAM & BAM files'. See the BAM file requirement in IGV documentation.
wget http://ccb.jhu.edu/software/tophat/downloads/test_data.tar.gz tar -xzf test_data.tar.gz # Assume required software are installed; e.g. through BDGE. export bdge_bowtie_PATH=/opt/RNA-Seq/bin/bowtie2-2.2.1 export bdge_tophat_PATH=/opt/RNA-Seq/bin/tophat-2.0.11.Linux_x86_64 export bdge_samtools_PATH=/opt/RNA-Seq/bin/samtools-0.1.19:/opt/RNA-Seq/bin/samtools-0.1.19/bcftools:/opt/RNA-Seq/bin/samtools-0.1.19/misc export PATH=$bdge_bowtie_PATH:$bdge_samtools_PATH:$bdge_tophat_PATH:$PATH tophat -r 20 test_ref reads_1.fq reads_2.fq # sort by location and index samtools sort tophat_out/accepted_hits.bam -o tophat_out_s.bam -@ 11 # Output tophat_out_s.bam samtools index tophat_out_s.bam # Output tophat_out_s.bam.bai
The "-r" option in tophat is related to mate-inner-dist. See the discussion here.
- Genome -> Load genome from file -> test_ref.fa
- File -> load from file -> tophat_out_s.bam (read_1.fq won't work)
- We can change the color of alignment by read strand by using the right click menu on *.bam window.
When we launch IGV, it is better to increase the default memory size (2GB). To do that, open igv.sh file and change -Xmx2000m to -Xmx4000m, for example to increase the memory from 2GB to 4GB.
- The reference genome (*.fa file) is shown on the bottom of IGV.
- Mouse over a sequence will show "Read name" (we can go back to fastq file to double check the sequence), "Location" (the location from mouse over), "Alignment start" (the start position).
- If there is a mismatch from a bp, it will be shown in a different color (T-red, C-blue, A-green, G-brown).
- The histogram-like plot is called Coverage.
- One sequence may be splitted after alignment.
- The <align_summary.txt> file (see below for its content) shows the number of reads in input and number of mapped reads with percentage of mapped.
- There are 142 reads shown on the IGV (verified by <align_summary.txt>).
The <test_ref.fa> file (reference genome) is shown below. The total length is 50 * 13 = 650 bp. We can verify this by looking at both the reference genome (bottom) and bp scale (top).
>test_chromosome AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA ACTACTATCTGACTAGACTGGAGGCGCTTGCGACTGAGCTAGGACGTGCC ACTACGGGGATGACGACTAGGACTACGGACGGACTTAGAGCGTCAGATGC AGCGACTGGACTATTTAGGACGATCGGACTGAGGAGGGCAGTAGGACGCT ACGTATTTGGCGCGCGGCGCTACGGCTGAGCGTCGAGCTTGCGATACGCC GTAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAG ACTATTACTTTATTATCTTACTCGGACGTAGACGGATCGGCAACGGGACT GTAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAG TTTTCTACTTGAGACTGGGATCGAGGCGGACTTTTTAGGACGGGACTTGC AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA AAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAAA
The <align_summary.txt> file is
Left reads:
Input : 100
Mapped : 72 (72.0% of input)
Right reads:
Input : 100
Mapped : 70 (70.0% of input)
71.0% overall read mapping rate.
Aligned pairs: 50
50.0% concordant pair alignment rate.
The first 2 reads (read length is 75) in <reads_1.fq> file looks like
@test_mRNA_150_290_0/1 TCCTAAAAAGTCCGCCTCGGTCTCAGTCTCAAGTAGAAAAAGTCCCGTTGGCGATCCGTCTACGTCCGAGTAAGA + IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII @test_mRNA_8_197_1/1 TCTGACTAGACTGGAGGCGCTTGCGACTGAGCTAGGACGTGACACTACGGGGATGGCGACTAGGACTACGGACGG + IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII
The first 2 reads in <reads_2.fq> file looks like
@test_mRNA_150_290_0/2 TACGTATTTGTCGCGCGGCCCTACGGCTGAGCGTCGAGCTTGCGATCCGCCACTATTACTTTATTATCTTACTCG + IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII @test_mRNA_8_197_1/2 GTATCGCAAGCTCGACGCTCAGCCGTAGGGCCGCGCGCCAAATACGTAGCGTCCTACTGCCCTCCTCAGTCCGAT + IIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIII
The IGV guide here shows how to interpret the colors for alignment data. For example, read bases that match are displayed in gray.
IGV Channel is a youtube channel where it contains some videos about the IGV.
Paire end sequencing
- http://www.illumina.com/technology/next-generation-sequencing/paired-end-sequencing_assay.html
- https://www.biostars.org/p/104522/
- http://www-huber.embl.de/users/anders/HTSeq/doc/count.html
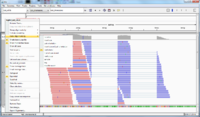
For paired end data, it is useful to use the right-click menu and select
- 'View as pairs' option. Pairs will be joined by a line.
- Color alignments by read strand.
The alignment of the right read works by 1) reverse the order and 2) apply compliment operator. For example, the following shows the reference sequence on the location of @test_mRNA_6_182_59/2. As we can see if we flip & apply the compliment of the sequence of @test_mRNA_6_182_59/2, they are almost matched except one base pair.
Reference GGACTATTTAGGACGATCGGACTGAGGAGGGCAGTAGGACGCTACGTATTTGGCGCGCGGCGCTACGGCTGAGCG @test_mRNA_6_182_59/2 CGCTCAGCCGTAGGGCCGCGCGCCAAATACGTAGCGTCCTACTGCCCTCCTCAGTCCGATCGTCCTAAATAGTCC
SRA000299 (single end)
This data was used in this post.
There are 4 libraries and 6 fastq files. The samples.txt is given below.
LibraryName LibraryLayout fastq1 fastq2 Kidney_0007_1.5pM SINGLE SRR002324.fastq Kidney_0007_3pM SINGLE SRR002320.fastq,SRR002325.fastq Liver_0007_1.5pM SINGLE SRR002322.fastq Liver_0007_3pM SINGLE SRR002321.fastq,SRR002323.fastq
Anders2013 (mix of single & paired end)
This is the data used in this wiki page. The sample meta data can be found at here.
Here we focus on a single-end library Untreated-6 which contains 2 fastq files (SRR031728.fastq, SRR031729.fastq).
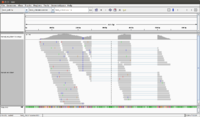
- In IGV, select the chromosome 2RHet and zoom in to the region of 1,638,560 bp. We are using the ref genome from ensembl.org as Anders2013 paper has used. Note the bp location number always starts at 1 for each chromosome. Here I am focusing on the read SRR031729.8202948 which has a color of a negative strand even the data is single end.
# Find the read sequence. First we find the line number using '-n' option of the 'grep' command brb@brb-T3500:~/Anders2013$ grep -n 8202948 SRR031729.fastq 32811789:@SRR031729.8202948 HWI-EAS299:3:65:192:596 length=75 32811791:+SRR031729.8202948 HWI-EAS299:3:65:192:596 length=75 # Next we display the text from line 32811789 to 32811790 brb@brb-T3500:~/Anders2013$ sed -n '32811789,32811790p' SRR031729.fastq @SRR031729.8202948 HWI-EAS299:3:65:192:596 length=75 CTTATCCTTTCTCTCTTGTATTTCCTGTGGAGGAAATTGACCTCAACCCATGGACTACCGAAACCTGGCAATATC
- Using an online DNA tool we can get the complement and inverse of the sequence (we only need to do this extra step for a negative strand.
GATATTGCCAGGTTTCGGTAGTCCATGGGTTGAGGTCAATTTCCTCCACAGGAAATACAAGAGAGAAAGGATAAG
- The reference genome shows the read starts at 2RHet:1638559, Cigar 75M. I write down the sequence by selecting 'Show all bases' in right-click menu. Not that the alignment is 100% for this read.
GATATTGCCAGGTTTCGGTAGTCCATGGGTTGAGGTCAATTTCCTCCACAGGAAATACAAGAGAGAAAGGATAAG
The ref genome seq matched with the complement & reverse seq of the (negative strand; shown in a purple color) read. This verifies the plot.
IGB
It is also a java-based software. Java version 1.8 is required.
- It provides similar functions as IGV.
- The memory handling is not as good as IGV (tested using Anders2013 data with the same 4000M setting).
- There are several tutorial videos for IGB and the user guide is good.
Other visualization tools
CHAPTER V : Visualizing next generation sequencing data
Count reads using htseq-count (sam -> count)
samples$countf = paste(samples$LibraryName, "count", sep=".")
gf = "Drosophila_melanogaster.BDGP5.70.gtf"
cmd = paste0("htseq-count -s no -a 10 ",
samples$LibraryName, "_sn.sam ",
gf," > ", samples$countf)
cmd
[1] "htseq-count -s no -a 10 Untreated-3_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > Untreated-3.count"
[2] "htseq-count -s no -a 10 Untreated-4_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > Untreated-4.count"
[3] "htseq-count -s no -a 10 CG8144_RNAi-3_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > CG8144_RNAi-3.count"
[4] "htseq-count -s no -a 10 CG8144_RNAi-4_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > CG8144_RNAi-4.count"
[5] "htseq-count -s no -a 10 Untreated-1_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > Untreated-1.count"
[6] "htseq-count -s no -a 10 CG8144_RNAi-1_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > CG8144_RNAi-1.count"
[7] "htseq-count -s no -a 10 Untreated-6_sn.sam Drosophila_melanogaster.BDGP5.70.gtf > Untreated-6.count"
Output from console
Lots of output
.... 37200000 SAM alignment records processed. 37300000 SAM alignment records processed. 37399340 SAM alignments processed.
-r pos option and 'Maximum alignment buffer size exceeded error'
If the input bam file is sorted by position, we can use -r pos option in HTSeq.scripts.count program. However, I got an error complaining Maximum alignment buffer size exceeded while pairing SAM alignments. Following the author's suggestion on the post, it is better to sort the bam file by read name and skip the -r option.
Performance
http://seqanswers.com/forums/showthread.php?t=18920 htseq-count was designed to use very little memory] (0.1GB from my observation). But the price is it is single thread and may takes several several hours. Note: Another tool is CuffLinks. See Sean Davis's tutorial.
Error: 'pair_alignments' needs a sequence of paired-end alignments
http://seqanswers.com/forums/showthread.php?t=41785 & https://www.biostars.org/p/111221/
Run the following to check if there are single-end in the bam files
samtools view -cF 1 bamfile
It it has, run the following to separate single-end and paired-end reads
samtools view -bf 1 foo.bam > foo.paired-end.bam samtools view -bF 1 foo.bam > foo.single-end.bam
Then run htseq-count separately and then combine the counts.
summarizeOverlaps() from GenomicAlignments package
Note that GenomicAlignments can read BAM files.
ReadCounter - htseq alternative
- it's multi threaded
- can produce the same result as htseq
- java based
featureCount from Subread
- Subread: high-performance read alignment, quantification and mutation discovery
- featureCounts: a ultrafast and accurate read summarization program
DESeq2 normalization
- Analyzing RNA-seq data with DESeq2
- DESeq2详细用法
- https://github.com/mgonzalezporta/TeachingMaterial/blob/master/doc/25.normalising.md
- http://www.sthda.com/english/wiki/rna-sequencing-data-analysis-counting-normalization-and-differential-expression
- Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2 by Love, Huber and Anders, 2014. Keywords: LFC (logarithmic fold change), shrinkage for dispersion estimation, MAP (maximum a posteriori), Shrinkage estimation of logarithmic fold changes, Wald test, Cook's distance for outlier detection. R packages: DESeq2, GenomicRanges and GenomicAlignments.
install.packages("~/Downloads/DESeq2paper_1.3.tar.gz", repos = NULL, type="source") source("http://bioconductor.org/biocLite.R") # https:// not work on my R 3.2.5 Ubuntu biocLite("DESeq2", ask=FALSE) # Got the following error because RcppArmidillo requires the following two libraries # /usr/bin/ld: cannot find -llapack # /usr/bin/ld: cannot find -lblas # Go to ubuntu software center search liblapack and libblas # select the following 6 components # libblas3gf, libblas-doc, libblas-dev, liblapack3gf, liblapack-doc, liblapack-dev # Then double check ls -l /usr/lib/liblapack* # ls -l /usr/lib/libblas* biocLite("GenomicRanges", ask=FALSE) biocLite("GenomicAlignments", ask=FALSE) install.packages("caret", repos="https://cran.rstudio.com")
- The count matrix and metadata, including the gene model and sample information, are stored in an S4 class derived from the SummarizedExperiment class of the GenomicRanges package. SummarizedExperiment objects containing count matrices can be easily generated using the summarizeOverlaps() function of the GenomicAlignments package.
- Log2 fold-change & DESeq2 model in a nutshell (youtube)
Input
DESeqDataSetFromHTSeqCount(sampleTable, directory, design) # sampleTable is like colData in the next 2 functions DESeqDataSetFromTximport(txi, colData, design) # txi is obtained by tximport() DESeqDataSetFromMatrix(countData, colData, design) # count data input DESeqDataSet(se, design) # e.g. the ''SummarizedExperiment'' object created by tximeta() # 'airway' package/object is an example
collapseReplicates
- StatQuest: RNA-seq - the problem with technical replicates
- Accounting for technical replicates in DESeq2
dds <- makeExampleDESeqDataSet(m=12)
# make data with two technical replicates for three samples
dds$sample <- factor(sample(paste0("sample",rep(1:9, c(2,1,1,2,1,1,2,1,1)))))
dds$run <- paste0("run",1:12)
ddsColl <- collapseReplicates(dds, dds$sample, dds$run)
# dds$sample is a grouping variable
# examine the colData and column names of the collapsed data
colData(ddsColl)
colnames(ddsColl)
# DataFrame with 9 rows and 4 columns
# condition sample run runsCollapsed
# <factor> <factor> <character> <character>
# sample1 A sample1 run6 run6,run9
# sample2 B sample2 run12 run12
# sample3 A sample3 run2 run2
# sample4 A sample4 run5 run5,run7
# sample5 A sample5 run1 run1
# sample6 A sample6 run4 run4
# sample7 B sample7 run8 run8,run10
# sample8 A sample8 run3 run3
# sample9 B sample9 run11 run11
# check that the sum of the counts for "sample1" is the same
# as the counts in the "sample1" column in ddsColl
matchFirstLevel <- dds$sample == levels(dds$sample)[1]
stopifnot(all(rowSums(counts(dds[,matchFirstLevel])) == counts(ddsColl[,1])))
Normalized counts = raw counts/sizeFactor
- How to access the normalized data of a DESeqDataSet
- How exactly DESeq2 correct for differences in sequencing depth and library composition to get normalized counts?
- To normalize for sequencing depth and RNA composition, DESeq2 uses the median of ratios method. On the user-end there is only one step, but on the back-end there are multiple steps involved. The algorithm performs multiple steps such as computing the pseudo-reference: geometric mean for each gene across all samples.
- StatQuest: DESeq2 part 1: Library Normalization (480p only)
- take the log of all the values (log0 = -Inf)
- average each row
- filter out genes with infinity
- subtract the average log value from the log(counts)
- calculate the median of the ratios for each sample
- convert the medians to "normal numbers" to get the final scaling factors for each sample (exp(these medians))
- divide the original read counts by the scaling factors
- Summary
- Calculate the normalization factor (size factor) for each sample:
- Create a pseudo-reference sample: For each gene, calculate the geometric mean of counts across all samples. This geometric mean serves as a pseudo-reference sample.
- Calculate the ratio of each sample to the reference: For each gene in each sample, divide the count by the geometric mean (pseudo-reference) calculated in step 1. This gives the ratio of each sample to the reference.
- For each sample, calculate the median of the ratios obtained in step 2. This median value is the size factor for that sample.
- Calculate the normalized count values: Divide the raw counts for each gene in each sample by the size factor for that sample. This gives the normalized count values.
- Calculate the normalization factor (size factor) for each sample:
- Differential expression analysis for sequence count data for DESeq package from Anders & Huber, 2010. If we want normalized counts, we need to run 1) estimateSizeFactors() for the normalization, and 2) counts(dds, normalized = TRUE) to extract normalized counts or normTransform(dds) on log2 scale. The normalization is part of the DESeq() function so you can extract counts after running that as well.
source("http://bioconductor.org/biocLite.R"); biocLite("pasilla") directory <- system.file("extdata", package="pasilla", mustWork=TRUE) sampleFiles <- grep("treated",list.files(directory),value=TRUE) sampleCondition <- sub("(.*treated).*","\\1",sampleFiles) sampleTable <- data.frame(sampleName = sampleFiles, fileName = sampleFiles, condition = sampleCondition) ddsHTSeq <- DESeqDataSetFromHTSeqCount(sampleTable = sampleTable, directory = directory, design= ~ condition) dds <- ddsHTSeq[ rowSums(counts(ddsHTSeq)) > 1, ] dds <- estimateSizeFactors(dds) filteredlog = counts(dds, normalized=T) filteredlog0 = counts(dds, normalized=F) filteredlog0[1:5, 1:5] # treated1fb.txt treated2fb.txt treated3fb.txt untreated1fb.txt untreated2fb.txt # FBgn0000008:003 0 1 0 1 1 # FBgn0000008:004 1 0 1 0 1 # FBgn0000008:005 4 1 1 2 2 # FBgn0000008:007 18 8 7 8 11 # FBgn0000008:008 4 1 0 0 1 scale(filteredlog0[1:5, ], center=F, scale=sizeFactors(dds))[, 1:5] # treated1fb.txt treated2fb.txt treated3fb.txt untreated1fb.txt untreated2fb.txt # FBgn0000008:003 0.0000000 1.246732 0.000000 0.9712273 0.6178182 # FBgn0000008:004 0.6815046 0.000000 1.093313 0.0000000 0.6178182 # FBgn0000008:005 2.7260184 1.246732 1.093313 1.9424546 1.2356364 # FBgn0000008:007 12.2670828 9.973855 7.653188 7.7698183 6.7960001 # FBgn0000008:008 2.7260184 1.246732 0.000000 0.0000000 0.6178182 filteredlog[1:5, 1:5] # treated1fb.txt treated2fb.txt treated3fb.txt untreated1fb.txt untreated2fb.txt # FBgn0000008:003 0.0000000 1.246732 0.000000 0.9712273 0.6178182 # FBgn0000008:004 0.6815046 0.000000 1.093313 0.0000000 0.6178182 # FBgn0000008:005 2.7260184 1.246732 1.093313 1.9424546 1.2356364 # FBgn0000008:007 12.2670828 9.973855 7.653188 7.7698183 6.7960001 # FBgn0000008:008 2.7260184 1.246732 0.000000 0.0000000 0.6178182# Retrieve raw counts and size factors raw_counts <- counts(dds) size_factors <- sizeFactors(dds) # Manually calculate normalized counts manual_normalized_counts <- sweep(raw_counts, 2, size_factors, FUN = "/") # Retrieve normalized counts from DESeq2 normalized_counts <- counts(dds, normalized = TRUE) # Compare the two matrices all.equal(manual_normalized_counts, normalized_counts)
High-Throughput Count Data
http://web.stanford.edu/class/bios221/book/Chap-CountData.html
estimateSizeFactorsForMatrix()
We can compare the output of estimateSizeFactorsForMatrix() with what you get by simply taking the column sums. See High-Throughput Count Data.
Use a robust regression to estimate the size factor instead of taking the sum of reads per sample.
The output estimateSizeFactorsForMatrix() is a vector of the number of samples.
estimateDispersions()
NB dispersion in DESeq2 correspond to Gamma-Poisson parametrisation?
dds <- makeExampleDESeqDataSet() # 1000 x 12 dds <- estimateSizeFactors(dds) dds <- estimateDispersions(dds) length(dispersions(dds)) # [1] 1000 plotDispEsts(dds) # x=mean of normalized counts, y=dispersion # plots the per-gene dispersion estimates together with # the fitted mean-dispersion relationship
So the output of estimateDispersions() is a vector of the number of genes.
- Two factors that we use to decide if we use shrinkage: high dispersion values, low gene counts
- Estimating Dispersion
DESeq() and gene filtering
- Remove genes with very low counts for all samples or let DESeq2 perform independent filtering? The safest threshold would be to not filter anything above row sum of 0... And our recommendation (and the default in DESeq2) is to let the genefilter software optimize the threshold, such that sensitivity (statistical power) is maximized.
- plotMA(), plotCounts
- DESeq: low p-values for genes with zero counts in all but one sample of the compared groups
Combine two DESeq2 objects (dds) for analysis
How to combine two DESeq2 objects (dds) for analysis
Designs and contrasts
- A guide to designs and contrasts in DESeq2
- How to correct for a covariate in DESeq2 that is linear with the condition of interest. Nested design.
- DESeq2 experimental design, 2 controls and 2 treated. Goal is to see how/if the two cell types respond differently to treatment.
- ~celltype + celltype:treatment
- Deseq2 design formula and analysis with internal controls. Goal is to compare two groups by taking the treatment into consideration.
- Factor1: group/cell-type
- Factor2: treatment
- DESeq2: Compare expression of one group to the mean of others
results(contrast)
- ?results
- Syntax: contrast=c('variable', 'numerator', 'denominator'). Example: contrast=c('Trt', 'Yes', 'No')
- Example 1: two conditions
dds <- makeExampleDESeqDataSet(m=4) dds <- DESeq(dds) res <- results(dds, contrast=c("condition","B","A")) # mean_B - mean_A res #log2 fold change (MLE): condition B vs A #Wald test p-value: condition B vs A #DataFrame with 1000 rows and 6 columns # baseMean log2FoldChange lfcSE stat pvalue padj # <numeric> <numeric> <numeric> <numeric> <numeric> <numeric> #gene1 2.18621 -3.029859 2.910105 -1.041151 0.297806 0.999351 #gene2 14.97050 -0.850521 1.112673 -0.764395 0.444632 0.999351 #gene3 9.31174 0.448988 1.379610 0.325446 0.744844 0.999351 #gene4 84.12136 0.333454 0.593804 0.561556 0.574419 0.999351 #gene5 21.52635 -0.538152 1.025702 -0.524667 0.599815 0.999351 counts(dds, normalized = T)[1:3, ] # sample1 sample2 sample3 sample4 #gene1 6.812072 0.9935038 0.00000 0.9392723 #gene2 11.677838 26.8246037 15.74392 5.6356338 #gene3 7.785225 7.9480307 18.69590 2.8178169 colData(dds) #DataFrame with 4 rows and 2 columns # condition sizeFactor # <factor> <numeric> #sample1 A 1.02759 #sample2 A 1.00654 #sample3 B 1.01627 #sample4 B 1.06465 # gene 1 > mean(c(6.81, .99)) # condition A [1] 3.9 > mean(c(0, 0.939)) # condition B [1] 0.4695 - Example 2: two conditions, one continuous variable (age)
set.seed(1234) dds <- makeExampleDESeqDataSet(n=100,m=12) dds$age <- rpois(12, 50) design(dds) <- ~ condition + age dds <- DESeq(dds) resultsNames(dds) # [1] "Intercept" "condition_B_vs_A" "age" results(dds) # OR results(dds, name="age") #log2 fold change (MLE): age #Wald test p-value: age #DataFrame with 100 rows and 6 columns # baseMean log2FoldChange lfcSE stat pvalue padj # <numeric> <numeric> <numeric> <numeric> <numeric> <numeric> #gene1 2.022210 -0.04620020 0.1445006 -0.3197232 0.749178 0.920829 #gene2 24.571867 0.00360519 0.0484505 0.0744097 0.940684 0.991664 #gene3 82.491501 0.00153923 0.0329532 0.0467095 0.962745 0.991664 results(dds, contrast=c("condition","B","A")) # OR results(dds, name="condition_B_vs_A") #log2 fold change (MLE): condition B vs A #Wald test p-value: condition B vs A #DataFrame with 100 rows and 6 columns # baseMean log2FoldChange lfcSE stat pvalue padj # <numeric> <numeric> <numeric> <numeric> <numeric> <numeric> #gene1 2.022210 3.0976656 1.854100 1.670711 0.0947788 0.910079 #gene2 24.571867 -0.1667077 0.635550 -0.262305 0.7930867 0.982211 #gene3 82.491501 0.2188290 0.431659 0.506948 0.6121910 0.982211 - Example 3: two conditions, two genotypes, with an interaction term
set.seed(1234) dds <- makeExampleDESeqDataSet(n=100,m=12) dds$genotype <- factor(rep(rep(c("I","II"),each=3),2)) design(dds) <- ~ genotype + condition + genotype:condition dds <- DESeq(dds) resultsNames(dds) # [1] "Intercept" "genotype_II_vs_I" # [3] "condition_B_vs_A" "genotypeII.conditionB" # the condition effect for genotype I (the main effect) results(dds, contrast=c("condition","B","A")) # the condition effect for genotype II # this is, by definition, the main effect *plus* the interaction term # (the extra condition effect in genotype II compared to genotype I). results(dds, list( c("condition_B_vs_A","genotypeII.conditionB") )) # the interaction term, answering: is the condition effect *different* across genotypes? results(dds, name="genotypeII.conditionB")
results(alpha, independentFiltering)
By default alpha=.1, Set independentFiltering=FALSE if we don't like independent filtering.
Negative binomial regression and returned from results()
A matrix with columns:
- baseMean: mean of normalized counts of all samples, normalizing for sequencing depth. It does not take into account gene length. The base mean is used in DESeq2 only for estimating the dispersion of a gene. See DESeq2 baseMean counts.
rowMeans(counts(dds, normalized=TRUE))
- log2FoldChange log2FoldChange calculation in DESeq2 output, 2019 STAT115 Lect7.4 DESeq2 by Shirley Liu. Theory behind DESeq2 & Null hypothesis. Introduction to DGE from hbctraining.
- [math]\displaystyle{ \begin{align} K_{ij} & \sim NB(\mu_{ij}, \alpha_i) \\ \mu_{ij} &= s_{j} q_{ij} \\ \log_2(q_{ij}) &= x_{j} \beta_i \end{align} }[/math]
[math]\displaystyle{ q_{ij} }[/math] is the normalized mean count for gene 𝑖 in sample 𝑗.
The coefficients [math]\displaystyle{ \beta_i }[/math] estimate give the log2 fold changes for gene i for each column of the model matrix [math]\displaystyle{ X }[/math].- In a simple linear regression [math]\displaystyle{ y = \beta_0 + \beta_1 x + \epsilon }[/math], if the covariate X takes only 0 or 1, we can show that the slope estimate [math]\displaystyle{ \frac{\sum(x_i-\bar{x})(y_i-\bar{y})}{\sum(x_i -\bar{x})^2} }[/math] can be simplified to the difference of group means [math]\displaystyle{ \bar{y}_1 - \bar{y}_0 }[/math]. This matches the idea of the fold change in gene expression.
- You can obtain standard log fold changes (no shrinkage) by using: DESeq(dds, betaPrior=FALSE)
- The null hypothesis being tested is: [math]\displaystyle{ H_0: \beta_i = 0 }[/math]. This tests whether the log fold change for the condition is zero, meaning there is no differential expression for gene 𝑖 between conditions.
- lfcSE standard error value (lfcSE) returned by DeSeq2
- Why the logFC is different between edgeR,DESeq2 results and fpkm logFC(log2(FPKM_tumor_mean/FPKM_normal_mean)? LogFCs are computed by negative binomial generalized linear models.
- Why my log2fcs are too high?
- stat: negative binomial GLM fitting for βi and Wald statistics by nbinomWaldTest(). log2Foldchange除以标准差所得
- pvalue & padj (Exploring DESeq2 results: Wald test from hbctraining)
Fold-change with continuous variables
how limma work with continous variables. It's the slope/fitted coefficient of the line.
Normalization-based vs log-ratio transformation-based methods
- Benchmarking differential expression analysis tools for RNA-Seq: normalization-based vs. log-ratio transformation-based methods
- If we use log2(normalized count + 1),有很低counts的基因将倾向于主导结果。作为一种解决方案,DESeq2为counts数据提供了stabilize the variance across the mean的转换。其中之一是regularized-logarithm transformation or rlog2。对于counts较高的基因,rlog转换可以得到与普通log2转换相似的结果。然而,对于counts较低的基因,所有样本的值都缩小到基因的平均值。See DESeq2详细用法 and the section Data transformations and visualization from the DESeq2 vignette.
Error with DESeq2
Error with DESeq2 “every gene contains at least one zero”
Source code
- counts(), normalizationFactors(), `normalizationFactor<-`, estimateSizeFactors(). DESeq2 -> R -> methods.R
- How can I extract normalized read count values from DESeq2 results?
? normalizationFactors
dds <- makeExampleDESeqDataSet(n=100, m=4)
normFactors <- matrix(runif(nrow(dds)*ncol(dds),0.5,1.5),
ncol=ncol(dds),nrow=nrow(dds),
dimnames=list(1:nrow(dds),1:ncol(dds)))
normFactors <- normFactors / exp(rowMeans(log(normFactors)))
normalizationFactors(dds) <- normFactors
dds <- DESeq(dds)
res = results(dds)
res[order(res$padj), ] %>% head
class(dds)
showMethods(class="DESeqDataSet") # a long list
isS4(assays(dds)) # [1] TRUE
slotNames(assays(dds))
# [1] "listData" "elementType" "elementMetadata" "metadata"
names(assays(dds))
# [1] "counts" "normalizationFactors" "mu" "H"
all(counts(dds) / normFactors == counts(dds, norm=T)) # TRUE
DESeq normalization 2010 ("median ratio")
edgeR normalization
library(baySeq) data(mobData) head(mobData) data(mobAnnotation) #?mobAnnotation head(mobAnnotation) library(edgeR) d <- DGEList(counts=mobData,group=factor(mobDataGroups)) d d.full <- d # keep the old one in case we mess up head(d.full$counts) head(cpm(d)) apply(d$counts, 2, sum) # total gene counts per sample d <- calcNormFactors(d) d
- ?calcNormFactors. If ‘object’ is a ‘matrix’, the output is a vector with length ‘ncol(object)’ giving the relative normalization factors.
y <- matrix( rpois(1000, lambda=5), nrow=200 ) calcNormFactors(y)
dge <- DGEList(M) dge <- calcNormFactors(dge) logCPM <- cpm(dge, log=TRUE) # section 2.16 of vignette
TMM, RLE, Upper quartile - A hypothetical situation
- Normalisation methods implemented in edgeR. The original code was modified a little bit.
hypothetical scenario:- The four samples have exactly the same depth (500 counts). However, since the case samples have half the number of transcripts than the controls (25 versus 50), the first 25 transcripts are sequenced at twice the depth.
- Only half of the transcripts are differentially expressed (the last 25 transcripts).
library(edgeR) control_1 <- rep(10, 50) control_2 <- rep(10, 50) case_1 <- c(rep(20, 25),rep(0,25)) case_2 <- c(rep(20, 25),rep(0,25)) eg1 <- data.frame( cont1 = control_1, cont2 = control_2, case1 = case_1, case2 = case_2 ) group <- rep(c('control', 'case'), each = 2) d <- DGEList(counts=eg1, group=group)No normalization:
d <- estimateCommonDisp(d) table(p.adjust(exactTest(d)$table$PValue, "BH") < .05) # TRUE # 50
TMM normalization.
- calcNormFactors() or normLibSizes() do not directly change your counts. Instead, it computes normalization factors in dge$samples$norm.factors.
d_tmm <- calcNormFactors(d, method="TMM") effective_lib_size <- d_tmm$samples$lib.size * d_tmm$samples$norm.factors effective_lib_size # [1] 353.5534 353.5534 707.1068 707.1068 t(t(d$counts) / effective_lib_size) |> head() # cont1 cont2 case1 case2 # 1 0.02828427 0.02828427 0.02828427 0.02828427 # 2 0.02828427 0.02828427 0.02828427 0.02828427 # 3 0.02828427 0.02828427 0.02828427 0.02828427 cpm(d_tmm, log = FALSE)/1e6 |> head(5) # cont1 cont2 case1 case2 # 1 0.02828427 0.02828427 0.02828427 0.02828427 # 2 0.02828427 0.02828427 0.02828427 0.02828427 # 3 0.02828427 0.02828427 0.02828427 0.02828427 d_tmm <- estimateCommonDisp(d_tmm) d_tmm <- exactTest(d_tmm) table(p.adjust(d_tmm$table$PValue, method="BH")<0.05) # FALSE TRUE # 25 25
- normLibSizes().
- See edgeR User's Guide.
DESeq2 for DE analysis
- By default, DESeq(test = "Wald"). Another test option is LRT.
- Vignette. Keyword: Wald test, coefficients [math]\displaystyle{ \beta }[/math], nbinomWaldTest. The coefficients βi give the log2 fold changes for gene i for each column of the model matrix X.
# Suppose the condition has 2 levels dds <- DESeqDataSetFromMatrix(countData = round(counts), colData = DataFrame(condition), design = ~ condition) dds <- DESeq(dds) res <- results(dds) res$stat # same as coef(dds)[, 2]/coef(dds, SE=T)[, 2] # coef(): extracting the matrix beta_{ir} for all genes i and model coefficients r # In the case of 2 conditions, the 1st column of coef() is intercept # and the 2nd column of coef() is the condition. all(res$stat == coef(dds)[, 2]/coef(dds, SE=T)[, 2], na.rm=T ) # [1] TRUE all(res$log2FoldChange == coef(dds)[, 2] ) all(res$lfcSE == coef(dds, SE=T)[, 2] ) - log2FoldChange represents the log normalized counts from the mean of level 2 vs level 1. But it is not clear how to calculate that exactly. See DESEq2 log2FoldChange and lfcSE calculations.
- According to ?results, The lfcSE gives the standard error of the log2FoldChange. For the Wald test, stat is the Wald statistic: the log2FoldChange divided by lfcSE, which is compared to a standard Normal distribution to generate a two-tailed pvalue.
- Differential expression analysis with DESeq2
- Videos from Chipster Tutorials https://www.youtube.com/watch?v=5tGCBW3_0IA
> samplesDESeq = with(samples, data.frame( shortname = I(shortname), countf = I(countf), condition = condition, LibraryLayout = LibraryLayout)) > samplesDESeq shortname countf condition LibraryLayout 1 CT.PA.1 Untreated-3.count CTL PAIRED 2 CT.PA.2 Untreated-4.count CTL PAIRED 3 KD.PA.3 CG8144_RNAi-3.count KD PAIRED 4 KD.PA.4 CG8144_RNAi-4.count KD PAIRED 5 CT.SI.5 Untreated-1.count CTL SINGLE 6 KD.SI.6 CG8144_RNAi-1.count KD SINGLE 7 CT.SI.7 Untreated-6.count CTL SINGLE > library("DESeq") cds <- newCountDataSetFromHTSeqCount(samplesDESeq) # quick, 15682 features, 7 samples cds <- estimateSizeFactors(cds) # quick sizeFactors(cds) # CT.PA.1 CT.PA.2 KD.PA.3 KD.PA.4 CT.SI.5 KD.SI.6 CT.SI.7 # 0.6991612 0.8104921 0.8217403 0.8941097 1.6431467 1.3720872 1.1041186 cds <- estimateDispersions(cds) # quick res <- nbinomTest(cds,"CTL","KD") # 44 seconds sum(res$pval <= .05, na.rm=T) # [1] 1574 sum(res$padj <= .05, na.rm=T) # [1] 730 options(width=100) res[1:3,] # id baseMean baseMeanA baseMeanB foldChange log2FoldChange pval padj # 1 FBgn0000003 0.000000 0.000000 0.000000 NaN NaN NA NA # 2 FBgn0000008 88.526695 88.486694 88.580029 1.001055 0.001520941 0.9298627 1 # 3 FBgn0000014 3.054418 2.327533 4.023599 1.728697 0.789684778 0.6839858 1
DESeqDataSet methods
- counts(): This method returns the raw count data for the genes in the DESeqDataSet object.
- colData(): This method returns the metadata (e.g., sample information) for the samples in the DESeqDataSet object.
- rowRanges()
- design()
- sizeFactors(): This method returns the size factors for the DESeqDataSet object, which are used to adjust for library size differences between samples.
- rld(): This method returns the regularized-logarithm (rlog) transformed data for the DESeqDataSet object, which is a variance-stabilizing transformation that can be used for visualization and clustering.
- plotDispEsts(): This method generates a plot of the dispersion estimates for the DESeqDataSet object, which can be used to diagnose the fit of the model.
- plotCounts(): This method generates a plot of the raw count data for the DESeqDataSet object, which can be used to visualize the distribution of the data.
- results(): This method returns the results of the differential expression analysis for the DESeqDataSet object, including the log2FoldChange values and the p-values.
- resultsNames()
- coef()
- plotMA(): MA plot
- Volcano plot:
- EnhancedVolcano::EnhancedVolcano(). Note:
- There are no default values for x and y parameters. All examples in the vignette use y = ‘pvalue’ (not ‘padj’).
- pCutoff means a p-value, not fdr adjustment
EnhancedVolcano(res, lab = rownames(res), pCutoff = 1e-05, FCcutoff = 1.0, x = 'log2FoldChange', y = 'pvalue')To draw adjusted p-values on the y-axis and use adjusted p-values as a cutoff (however, the label for some sig genes are missing. IOW, ggplot2 solution is better)
EnhancedVolcano(res, lab = rownames(res), pCutoff = 0.05, pCutoffCol = "padj", FCcutoff = 1.0, ylim = c(0, max(-log10(res$padj), na.rm = TRUE)), labSize = 2.5, x = 'log2FoldChange', y = 'padj', title = 'Volcano plot', ylab = bquote(~-Log[10]~'Adjusted p-value'), legendLabels = c('NS', 'Log2 FC', 'Adjusted p-value', 'Adjusted p-value & Log2 FC')) - ggplot2
ggplot(res, aes(x = log2FoldChange, y = -log10(pvalue))) + geom_point() + xlab("log2 Fold Change") + ylab("-log10(p-value)") + ggtitle("Volcano Plot")If we like to add vertical and horizontal cutoff lines and change to use adjusted p-values (padj column), we can follow Making Volcano Plots With ggplot2:
library(ggplot2) library(ggrepel) data <- res data$gene <- rownames(res) ggplot(data, aes(x = log2FoldChange, y = -log10(padj))) + geom_point(aes(color = padj < 0.05 & abs(log2FoldChange) > 1), alpha = 0.5) + scale_color_manual(values = c("black", "red"), na.translate = F) + geom_vline( xintercept = c(-1, 1), linetype = 'dashed' ) + geom_hline( yintercept = -log10(.05), linetype = 'dashed' ) + theme_bw() + labs(title = "Volcano Plot", x = "Log2 Fold Change", y = "-Log10 Adjusted P-Value") + geom_label_repel( data = significant_genes, aes(label = gene), size=3, box.padding = 0.25, # default point.padding = 1e-06, # default max.overlaps = 10 # default )
- EnhancedVolcano::EnhancedVolcano(). Note:
Pre-filter
- DESeq2 vignette. row sum >= 10.
- Modeling and cleaning RNA-seq data significantly improve detection of differentially expressed genes 2022.
betaPrior is FALSE by default
Different results in DESeq2 when using betaPrior=TRUE and betaPrior=FALSE
lfcShrink
Getting NAs in p-values
Getting NAs in both p-values (normal and adjusted) in DEseq2
Comparison
A comparison study on modeling of clustered and overdispersed count data for multiple comparisons Kruppa 2020
Wald vs Likelihood ratio test
- HELP - DESeq2 difference between LRT and Wald Test in RNA-seq/ATAC-seq.
- The Wald tests are specific to a given contrast. You are specifically asking if there is evidence that the coefficient comparing two groups is larger than zero... I would imagine that the vast majority of people use Wald tests exclusively, as they are mostly wanting to make specific comparisons.
- If you fit a full and reduced model and then compute a likelihood ratio test, you are testing if the full model fits the data better than the reduced model. It's an omnibus test that tells you that at least one coefficient is significant, much like an F-test in a conventional linear regression.
DESeq2, edgeR, limma
- Genome ⟶ edgeR vs DESeq2 vs limma
- 差异分析三巨头 —— DESeq2、edgeR 和 limma 包 | 附完整代码 + 注释
- How do I explain the difference between edgeR, LIMMA, DESeq etc. to experimental Biologist/non-bioinformatician.
- DESeq and EdgeR are very similar and both assume that no genes are differentially expressed.
- Limma / Voom is different in that it normalises via the very successful (for microarrays) quantile nomalisation, where an attempt is made to match gene count distributions across samples in your dataset.
- Differential Expression with Limma-Voom
- RNA Seq Differential Analyses Guideline in R
DESeq2 paired multifactor test
DESeq2 paired multifactor test
t-test vs DESeq2
FPKM t-test vs DEseq2. t-tests are parametric, based on the normal distribution. t-tests are expected to yield far fewer DEGs simply because at low sample size they're massively underpowered, please read papers to learn why. That is the basis why methods such as DESeq2 even exist. RNA-seq is not normally distributed, that is well-known.
Time series
- RNA-seq workflow: gene-level exploratory analysis and differential expression
- Note for DEseq2 time course analysis, Note 2
Interaction
Complex DESeq2 Experimental Design
Should I subset the data for analyses
If I have multiple groups, should I run all together or split into pairs of groups?
File Format
- http://genome.ucsc.edu/FAQ/FAQformat.html which includes a lot of common formats like BAM, BED, bedGraph, bigBed, bigWig, VCF, WIG, et al.
- http://stephenturner.us/biofiletypes/
fastq
The wikipedia website provides information to convert FASTQ to FASTA format (when do we want to do that?).
fastq format
A FASTQ file normally uses four lines per sequence.
@SEQ_ID GATTTGGGGTTCAAAGCAGTATCGATCAAATAGTAAATCCATTTGTTCAACTCACAGTTT + !''*((((***+))%%%++)(%%%%).1***-+*''))**55CCF>>>>>>CCCCCCC65
- Line 1 begins with a '@' character and is followed by a sequence identifier and an optional description (like a FASTA title line).
- Line 2 is the raw sequence letters.
- Line 3 begins with a '+' character and is optionally followed by the same sequence identifier (and any description) again.
- Line 4 encodes the quality values for the sequence in Line 2, and must contain the same number of symbols as letters in the sequence.
It seems line 3 & 4 are not required. See the simulated fastq files on http://genomespot.blogspot.com/2014/11/dna-aligner-accuracy-bwa-bowtie-soap.html. I can use BWA to run alignment without a problem:)
Paired end
Forward and reverse reads in paired-end sequencing
The paired files can be two formats. See Bioinformatics at COMAV
Format1:
Fastq file 1 @molecule_1 1st_read_from_pair @molecule_2 1st_read_from_pair @molecule_3 1st_read_from_pair Fastq file 2 @molecule_1 2nd_read_from_pair @molecule_2 2nd_read_from_pair @molecule_3 2nd_read_from_pair
Format 2:
Interleaved Fastq file @molecule_1 1st_read_from_pair @molecule_1 2nd_read_from_pair @molecule_2 1st_read_from_pair @molecule_2 2nd_read_from_pair @molecule_3 1st_read_from_pair @molecule_3 2nd_read_from_pair
Note from my experiment, if we flip the two fastq files order in the BWA command, the resulting bam file will diff at FLAG (2nd) column. The final vcf file is not changed.
FastQValidator
http://genome.sph.umich.edu/wiki/FastQValidator
fasta
FASTA format is a text-based format for representing nucleotide sequences . cf. FASTQ files provide more information than FASTA since FASTQ format stores both a biological sequence (usually nucleotide sequence) and its corresponding quality scores.
The reference genome is saved in FASTA format.
Normally the first line in a FASTA file starts with a ">" (greater-than) symbol. Subsequent lines starting with a semicolon would be ignored by software. Following the initial line (used for a unique description of the sequence) is the actual sequence itself in standard one-letter code. A multiple sequence FASTA format would be obtained by concatenating several single sequence FASTA files.
For the fruit fly genome file Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa. The grep -n command can be used to get the row numbers of some keyword (chromosome names in this case).
$ head Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa >2L dna:chromosome chromosome:BDGP5:2L:1:23011544:1 REF CGACAATGCACGACAGAGGAAGCAGAACAGATATTTAGATTGCCTCTCATTTTCTCTCCC ATATTATAGGGAGAAATATGATCGCGTATGCGAGAGTAGTGCCAACATATTGTGCTCTTT GATTTTTTGGCAACCCAAAATGGTGGCGGATGAACGAGATGATAATATATTCAAGTTGCC GCTAATCAGAAATAAATTCATTGCAACGTTAAATACAGCACAATATATGATCGCGTATGC GAGAGTAGTGCCAACATATTGTGCTAATGAGTGCCTCTCGTTCTCTGTCTTATATTACCG CAAACCCAAAAAGACAATACACGACAGAGAGAGAGAGCAGCGGAGATATTTAGATTGCCT ATTAAATATGATCGCGTATGCGAGAGTAGTGCCAACATATTGTGCTCTCTATATAATGAC TGCCTCTCATTCTGTCTTATTTTACCGCAAACCCAAATCGACAATGCACGACAGAGGAAG CAGAACAGATATTTAGATTGCCTCTCATTTTCTCTCCCATATTATAGGGAGAAATATGAT $ grep -n ">" Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa 1:>2L dna:chromosome chromosome:BDGP5:2L:1:23011544:1 REF 383528:>2LHet dna:chromosome chromosome:BDGP5:2LHet:1:368872:1 REF 389677:>2R dna:chromosome chromosome:BDGP5:2R:1:21146708:1 REF 742124:>2RHet dna:chromosome chromosome:BDGP5:2RHet:1:3288761:1 REF 796938:>3L dna:chromosome chromosome:BDGP5:3L:1:24543557:1 REF 1205999:>3LHet dna:chromosome chromosome:BDGP5:3LHet:1:2555491:1 REF 1248592:>3R dna:chromosome chromosome:BDGP5:3R:1:27905053:1 REF 1713678:>3RHet dna:chromosome chromosome:BDGP5:3RHet:1:2517507:1 REF 1755638:>4 dna:chromosome chromosome:BDGP5:4:1:1351857:1 REF 1778170:>U dna:chromosome chromosome:BDGP5:U:1:10049037:1 REF 1945655:>Uextra dna:chromosome chromosome:BDGP5:Uextra:1:29004656:1 REF 2429067:>X dna:chromosome chromosome:BDGP5:X:1:22422827:1 REF 2802782:>XHet dna:chromosome chromosome:BDGP5:XHet:1:204112:1 REF 2806185:>YHet dna:chromosome chromosome:BDGP5:YHet:1:347038:1 REF 2811970:>dmel_mitochondrion_genome dna:chromosome chromosome:BDGP5:dmel_mitochondrion_genome:1:19517:1 REF $ wc -l *.fa 2812296 Drosophila_melanogaster.BDGP5.70.dna.toplevel.fa
For human,
$ grep -n ">" ~/igenomes/Homo_sapiens/UCSC/hg19/Sequence/WholeGenomeFasta/genome.fa 1:>chrM 334:>chr1 4985348:>chr2 9849337:>chr3 13809787:>chr4 17632874:>chr5 21251181:>chr6 24673484:>chr7 27856259:>chr8 30783541:>chr9 33607811:>chr10 36318507:>chr11 39018639:>chr12 41695678:>chr13 43999077:>chr14 46146069:>chr15 48196698:>chr16 50003795:>chr17 51627701:>chr18 53189247:>chr19 54371828:>chr20 55632340:>chr21 56594939:>chr22 57621032:>chrX 60726445:>chrY
chromosome names
When I apply the above method to examine the genome.fa files from Ensembl, NCBI and UCSC using the command
grep -n ">" genome.fa | cut -d' ' -f1 | cut -d '>' -f2 # for example grep -n ">" ~/Downloads/Homo_sapiens.GRCh38.dna.chromosome.22.fa | cut -d' ' -f1 | cut -d '>' -f2 # 22
I get the following result
- Ensembl:
- GRCh37: 1, 2, 3, ...., X, Y, MT
- GRCh38: 1, 2, 3, ... (see note below)
- NCBI:
- build37.1, build37.2: 1, 2, 3, ....,X, Y, MT
- GRCh38: chr1, chr2, ...., chr22, chrX, chrY, chrM, chr1_KI270706v1_random, ....., chrEBV
- UCSC:
- hg18, hg19: chr1, chr2, ...., chr22, chrX, chrY, chrM
- hg38: chr1, chr2, ..., chr22, chrX, chrY, chrM, ..., chrUn_GL000218v1, chrEBV
Note that
- Neither Illumina or Biowulf provide GRCh38 from Ensembl (maybe for a good reason). Ensembl (http://ftp.ensembl.org/pub/release-76/fasta/homo_sapiens/dna) does provide GRCh38.
- GRCh38 from Ensembl is still following its GRCh37 convention; it won't include 'chr' on their chromosome names. That is, even the same name GRCh38 was used in both Ensembl and NCBI, they have different conventions.
chromosome sizes
https://www.biostars.org/p/134807/
GENOMEFA=Arabidopsis_thaliana.TAIR10.23.dna.genome.fa # Method 1: use pyfaidx # upgrade pip from 8.1.2 to 9.0.1 sudo pip install --upgrade pip sudo pip install pyfaidx $ cd /tmp $ faidx $GENOMEFA -i chromsizes > sizes.genome $ cat sizes.genome Pt 154478 Mt 366924 4 18585056 2 19698289 3 23459830 5 26975502 1 30427671 # Method 2: use samtools $ /opt/SeqTools/bin/samtools-1.3/samtools faidx $GENOMEFA $ cat $GENOMEFA.fai Pt 154478 51 60 61 Mt 366924 157155 60 61 4 18585056 530246 60 61 2 19698289 19425104 60 61 3 23459830 39451749 60 61 5 26975502 63302628 60 61 1 30427671 90727773 60 61 $ cut -f1,2 $GENOMEFA.fai > sizes.genome $ cat sizes.genome2 Pt 154478 Mt 366924 4 18585056 2 19698289 3 23459830 5 26975502 1 30427671
R approach
library(Biostrings)
x <- readDNAStringSet("~/igenomes/Homo_sapiens/UCSC/hg38/Sequence/WholeGenomeFasta/genome.fa")
y <- width(x[1:24])
barplot(y, las = 2)
y
# chr1 chr2 chr3 chr4 chr5 chr6 chr7 chr8
# 248956422 242193529 198295559 190214555 181538259 170805979 159345973 145138636
# chr9 chr10 chr11 chr12 chr13 chr14 chr15 chr16
# 138394717 133797422 135086622 133275309 114364328 107043718 101991189 90338345
# chr17 chr18 chr19 chr20 chr21 chr22 chrX chrY
# 83257441 80373285 58617616 64444167 46709983 50818468 156040895 57227415
Remove/Add 'chr' from fasta
sed 's/chr//g' ucsc.hg19.fa >ucsc.hg19.nochr.fa # or cat hg19.fa | sed 's/>chr/>/g' > hg19_new.fa
Similarly to add 'chr' to fasta:
cat hg19.fa | sed 's/>/>chr/g' > hg19_new.fa
Convert Fasta to Fastq
It is OK to use Fasta file as the input sequencing files in alignment programs.
If we still want to convert FASTA to FASTQ, two methods are given with an example here. Note quality scores are padded.
Convert Fastq to Fasta
Note the quality scores are discarded.
- https://wikis.utexas.edu/display/bioiteam/Mapping+tutorial
- https://www.biostars.org/p/85929/. Use cat test.fastq | paste - - - - | sed 's/^@/>/g'| cut -f1-2 | tr '\t' '\n' .
$ cat test.fastq @SRR002051.3554692 :8:290:671:993 length=33 CCATTCCCCAAAACAGTAAAACCACACACCGAG +SRR002051.3554692 :8:290:671:993 length=33 40&&+(&#$&3""%$#"#%$)%"%"%$/%%""# @SRR002051.3554693 :8:290:941:550 length=33 ACTAACATCCCATAATCGTACCTCCTACCATCA +SRR002051.3554693 :8:290:941:550 length=33 IIII*I$+(/II'$&:I"%""""&"&$%"'""I $ cat test.fastq | paste - - - - | sed 's/^@/>/g'| cut -f1-2 | tr '\t' '\n' >SRR002051.3554692 :8:290:671:993 length=33 CCATTCCCCAAAACAGTAAAACCACACACCGAG >SRR002051.3554693 :8:290:941:550 length=33 ACTAACATCCCATAATCGTACCTCCTACCATCA
- Convert fastq files to two fasta files with one containing the reads and another containing the quality scores. In the example below we try to separate paired end reads generated by BEERS program.
$ head simulated_reads_testbeers.fa
>seq.1a
CGAAGAAGGACCCAAAGATGACAAGGCTCACAAAGTACACCCAGGGCAGTTCATACCCCATGGCATCTTGCATCCAGTAGAGCACATCGGTCCAGCCTTC
>seq.1b
GCTCGAGCTGTTCCTTGGACGAATGCACAAGACGTGCTACTTCCTGGGATCCGACATGGAAGCGGAGGAGGACCCATCGCCCTGTGCATCTTCGGGATCA
>seq.2a
GCCCCAGCAGAGCCGGGTAAAGATCAGGAGGGTTAGAAAAAATCAGCGCTTCCTCTTCCTCCAAGGCAGCCAGACTCTTTAACAGGTCCGGAGGAAGCAG
>seq.2b
ATGAAGCCTTTTCCCATGGAGCCATATAACCATAATCCCTCAGAAGTCAAGGTCCCAGAATTCTACTGGGATTCTTCCTACAGCATGGCTGATAACAGAT
>seq.3a
CCCCAGAGGAGCGCCACCTGTCCAAGATGCAGCAGAACGGCTACGAAAATCCAACCTACAAGTTCTTTGAGCAGATGCAGAACTAGACCCCCGCCACAGC
$ awk 'NR % 4 == 1 {print ">" $0 } NR % 4 == 2 {print $0}' simulated_reads_testbeers.fa > my.fasta
$ head my.fasta
>>seq.1a
CGAAGAAGGACCCAAAGATGACAAGGCTCACAAAGTACACCCAGGGCAGTTCATACCCCATGGCATCTTGCATCCAGTAGAGCACATCGGTCCAGCCTTC
>>seq.2a
GCCCCAGCAGAGCCGGGTAAAGATCAGGAGGGTTAGAAAAAATCAGCGCTTCCTCTTCCTCCAAGGCAGCCAGACTCTTTAACAGGTCCGGAGGAAGCAG
>>seq.3a
CCCCAGAGGAGCGCCACCTGTCCAAGATGCAGCAGAACGGCTACGAAAATCCAACCTACAAGTTCTTTGAGCAGATGCAGAACTAGACCCCCGCCACAGC
>>seq.4a
CGGGTGGAGGCTGGCCAAGCAACGAAGGAGATAAAATTGCAGCTGGCATTGTGTGTGCGCGCGCCCGCGGCGGCGGCCAGCAGGGGTGCGCGCGGAGCGG
>>seq.5a
CGCTCACCTTCAATGTCACTGGCAAACATACTGCCTACCAGCTACTCCTCAGCCTTACACCTTAAGCATACGTCCTGCTCTTACTCTTCGAGTTGAGGGT
$ awk 'NR % 4 == 0 {print ">" $0 } NR % 4 == 3 {print $0}' simulated_reads_testbeers.fa > my.fasta
$ head my.fasta
>seq.1b
>GCTCGAGCTGTTCCTTGGACGAATGCACAAGACGTGCTACTTCCTGGGATCCGACATGGAAGCGGAGGAGGACCCATCGCCCTGTGCATCTTCGGGATCA
>seq.2b
>ATGAAGCCTTTTCCCATGGAGCCATATAACCATAATCCCTCAGAAGTCAAGGTCCCAGAATTCTACTGGGATTCTTCCTACAGCATGGCTGATAACAGAT
>seq.3b
>GTTTGATACAGCTAAATTCTTTACAGTACACAAAACCCATTAATAATGTAGTATAGAGACCAAAATGTAAAGAGAGATAGAATACATTACTGATGTGTGG
>seq.4b
>CGGGGCCCCGCCCGCGGCTAGGGAGGTGGCCGCCCTGGCCCGAGCCTGCTGCCCACGGCCGCGGCACTCAGCTCGCACTCCTTCAGCCCGGGAGGCCGGG
>seq.5b
>ACTCCCTGATAGTCTATGGAAGGAAAATGACAACTATTTTAGAATATTTCTAGTTTGTTTTTTCAGTGATCTTTTCATCCAGGCCTTGTTACTGTTACAG
- https://gif.biotech.iastate.edu/converting-fastq-fasta
- Bioconductor packages Biostrings or shortread https://support.bioconductor.org/p/38338/
GATK
When I use Ensembl/GRCh37 as the reference to run GATK, I got an error
##### ERROR MESSAGE: Input files /home/brb/SeqTestdata/usefulvcf/hg19/gatk/common_all_20160601.vcf and reference have incompatible contigs. Please see http://gatkforums.broadinstitute.org/discussion/63/input-files-have-incompatible-contigsfor more information. Error details: No overlapping contigs found. ##### ERROR /home/brb/SeqTestdata/usefulvcf/hg19/gatk/common_all_20160601.vcf contigs = [chr1, chr2, chr3, chr4, chr5, chr6, chr7, chr8, chr9, chr10, chr11, chr12, chr13, chr14, chr15, chr16, chr17, chr18, chr19, chr20, chr21, chr22, chrX, chrY] ##### ERROR reference contigs = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, X, Y, MT]
The conclusion is I need to check if the chromosome name in dbsnp vcf file (ftp://ftp.ncbi.nih.gov/snp/organisms/) and reference fa file has the same naming convention.
For example, for Ensemble/GRCh37, I should NOT use the GATK version (i.e. no 'chr' was added) from ftp://ftp.ncbi.nlm.nih.gov/snp/organisms/human_9606_b147_GRCh37p13/VCF/
(P.S. For UCSC hg19, we shall use the GATK version ftp://ftp.ncbi.nlm.nih.gov/snp/organisms/human_9606_b147_GRCh37p13/VCF/GATK/)
But for UCSC/hg38, I should use the GATK version ('chr' was added) from ftp://ftp.ncbi.nlm.nih.gov/snp/organisms/human_9606_b147_GRCh38p2/VCF/GATK/. Read the readme
So I end up with the file structure in my PC
- usefulvcf/hg19/common_all*.vcf (no 'chr' was added)
usefulvcf/hg38/gatk/common_all*.vcfusefulvcf/hg38/chr/common_all*.vcf ('chr' was added)
gff/gtf
- Comparing Ensembl GTF and cDNA/ensembl's Fasta and GTF annotation files do not match
- The GTF file can be downloaded from the Tophat website. After that, extract the file genes.gtf according to the instruction in BDGE website.
- GTF file can also be downloaded from
- UCSC Genome Bioinformatics or UCSC Table Browser. See the Mapping reads to the genome tutorial in homer.salk.edu.
- NCBI
- Ensembl
- GTF file can change. For example there are 7 versions of hg19 reference genome,
brb@T3600 ~ $ ls -l ~/igenomes/Homo_sapiens/UCSC/hg19/Annotation/ total 4 drwxrwxr-x 9 brb brb 4096 Jul 24 2015 Archives lrwxrwxrwx 1 brb brb 30 Mar 10 03:03 Genes -> Archives/archive-current/Genes lrwxrwxrwx 1 brb brb 35 Mar 10 03:03 README.txt -> Archives/archive-current/README.txt lrwxrwxrwx 1 brb brb 33 Mar 10 03:03 SmallRNA -> Archives/archive-current/SmallRNA lrwxrwxrwx 1 brb brb 34 Mar 10 03:03 Variation -> Archives/archive-current/Variation brb@T3600 ~ $ ls -lH ~/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Archives total 28 drwxrwxr-x 4 brb brb 4096 Mar 15 2012 archive-2010-09-27-22-25-17 drwxrwxr-x 5 brb brb 4096 Mar 15 2012 archive-2011-01-27-18-25-49 drwxrwxr-x 5 brb brb 4096 Mar 15 2012 archive-2011-08-30-21-45-18 drwxrwxr-x 5 brb brb 4096 Mar 15 2012 archive-2012-03-09-03-24-41 drwxrwxr-x 5 brb brb 4096 Oct 1 2013 archive-2013-03-06-11-23-03 drwxrwxr-x 5 brb brb 4096 Jun 2 2014 archive-2014-06-02-13-47-56 drwxrwxr-x 5 brb brb 4096 Jul 24 2015 archive-2015-07-17-14-32-32 lrwxrwxrwx 1 brb brb 27 Mar 10 03:35 archive-current -> archive-2015-07-17-14-32-32
- GTF file is used in RNA-Seq reads alignment and count reads.
- Tophat - If this option is provided, TopHat will first extract the transcript sequences and use Bowtie to align reads to this virtual transcriptome first. Only the reads that do not fully map to the transcriptome will then be mapped on the genome. The reads that did map on the transcriptome will be converted to genomic mappings (spliced as needed) and merged with the novel mappings and junctions in the final tophat output.
- STAR - GTF file (containing annotated transcripts) can be used in creating STAR index files (--sjdbGTFfile option in STAR --runMode genomeGenerate). STAR will extract splice junctions from this file and use them to greatly improve accuracy of the mapping. Note that in the mapping step there is no need to supply the GTF file again.
- htseq-count: one of two required inputs (the other one is a sorted bam file).
- The meaning of Gene isoform in wikipedia. It contains links to TSS, CDS, UTR.
- The meaning of transcriptome in wikipedia. Each row in a GTF file contains one feature and the the feature's transcript id and gene name. See gist.
- Gene name in GTF file
$ grep KRAS ~/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf | wc -l 23
- What is CDS in GTF file? CDS (contiguous sequence) CDS is the sequence that actually makes proteins. See biostars.org.
- What is exon? A region of the transcript sequence within a gene which is not removed from the primary RNA transcript by RNA splicing. See its definition on
- GFF3 format from sequenceontology.org
The following output shows how many features from different sources.
brb@brb-T3500:/tmp$ wc -l ~/igenomes/Homo_sapiens/UCSC/hg19/Sequence/WholeGenomeFasta/genome.fa 61913917 /home/brb/igenomes/Homo_sapiens/UCSC/hg19/Sequence/WholeGenomeFasta/genome.fa brb@brb-T3500:/tmp$ wc -l ~/igenomes/Homo_sapiens/NCBI/build37.2/Sequence/WholeGenomeFasta/genome.fa 44224234 /home/brb/igenomes/Homo_sapiens/NCBI/build37.2/Sequence/WholeGenomeFasta/genome.fa brb@brb-T3500:/tmp$ wc -l ~/igenomes/Homo_sapiens/Ensembl/GRCh37/Sequence/WholeGenomeFasta/genome.fa 51594937 /home/brb/igenomes/Homo_sapiens/Ensembl/GRCh37/Sequence/WholeGenomeFasta/genome.fa brb@brb-T3500:~/igenomes/Homo_sapiens$ wc -l UCSC/hg19/Annotation/Genes/genes.gtf 869204 UCSC/hg19/Annotation/Genes/genes.gtf brb@brb-T3500:~/igenomes/Homo_sapiens$ wc -l NCBI/build37.2/Annotation/Genes/genes.gtf 819119 NCBI/build37.2/Annotation/Genes/genes.gtf brb@brb-T3500:~/igenomes/Homo_sapiens$ wc -l Ensembl/GRCh37/Annotation/Genes/genes.gtf 2280612 Ensembl/GRCh37/Annotation/Genes/genes.gtf
An example from UCSC/hg19. As you can see from IGV and the output of the first few rows,
- the locations of each rows are not exclusive. For example, rows #3 and #4 are overlapped that matches with what we saw on IGV (fly example)
- one row represents one feature; one transcript consists several features
- one gene (e.g. WASH7P) can be split over multiple regions (UCSC/hg19 example).
$ more ~/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf chr1 unknown exon 11874 12227 . + . gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS16932"; chr1 unknown exon 12613 12721 . + . gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS16932"; chr1 unknown exon 13221 14409 . + . gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS16932"; chr1 unknown exon 14362 14829 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 14970 15038 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 15796 15947 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 16607 16765 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 16858 17055 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 17233 17368 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 17369 17436 . - . gene_id "MIR6859-3"; gene_name "MIR6859-3"; transcript_id "NR_107063_2"; tss_id "TSS25677"; chr1 unknown exon 17369 17436 . - . gene_id "MIR6859-2"; gene_name "MIR6859-2"; transcript_id "NR_107062_1"; tss_id "TSS25677"; chr1 unknown exon 17369 17436 . - . gene_id "MIR6859-4"; gene_name "MIR6859-4"; transcript_id "NR_128720_2"; tss_id "TSS25677"; chr1 unknown exon 17369 17436 . - . gene_id "MIR6859-1"; gene_name "MIR6859-1"; transcript_id "NR_106918_2"; tss_id "TSS25677"; chr1 unknown exon 17606 17742 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 17915 18061 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 18268 18366 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 24738 24891 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 29321 29370 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; chr1 unknown exon 30366 30503 . + . gene_id "MIR1302-11"; gene_name "MIR1302-11"; transcript_id "NR_036268"; tss_id "TSS10004";
The following table gives an example (UCSC/hg19) from one feature. Notice that the last column contains transcript ID, tss ID and gene ID. If the GTF is obtained from Ensembl, this column also has exon ID.
| 1 | seqname | chr1 (called 1 from NCBI & Ensembl) |
| 2 | source | unknown |
| 3 | feature | exon/transcript |
| 4 | start | 11874 |
| 5 | end | 12227 |
| 6 | score | . |
| 7 | strand | + |
| 8 | frame | 0 |
| 9 | attribute | gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS14844"; |
The gene attributes (gene_id, transcript_id, exon_number, gene_name, gene_biotype, transcript_name, protein_id) appears on the mouse-over pop-up in IGV. See the 1st screenshot below from fly genome (imported manually by myself to IGV). From the 1st screenshot it is clear that one position may contain more than one transcripts.
This post on seqanswers.com has a discussion about the GTF file, exon, intron, CDS, exon, etc. It says
"Exon" refers to transcription (DNA->RNA) and "CDS" to translation (RNA->Proteins). These are two different biological mechanisms.
rtracklayer package
See also the vignette of SingscoreAMLMutations
brb@brb-T3500:~/igenomes/Homo_sapiens$ R
> x <- read.delim("~/igenomes/Homo_sapiens/Ensembl/GRCh37/Annotation/Genes/genes.gtf", header=FALSE, as.is=TRUE)
> dim(x)
[1] 2280612 9
> x[1:2, 1:8]
V1 V2 V3 V4 V5 V6 V7 V8
1 1 processed_transcript exon 11869 12227 . + .
2 1 processed_transcript transcript 11869 14409 . + .
> strsplit(x[1:2, 9], ";") # not the same number of elements so we cannot apply as.data.frame()
[[1]]
[1] "exon_id ENSE00002234944" " exon_number 1"
[3] " gene_biotype pseudogene" " gene_id ENSG00000223972"
[5] " gene_name DDX11L1" " gene_source ensembl_havana"
[7] " transcript_id ENST00000456328" " transcript_name DDX11L1-002"
[9] " transcript_source havana" " tss_id TSS15145"
[[2]]
[1] "gene_biotype pseudogene" " gene_id ENSG00000223972"
[3] " gene_name DDX11L1" " gene_source ensembl_havana"
[5] " transcript_id ENST00000456328" " transcript_name DDX11L1-002"
[7] " transcript_source havana" " tss_id TSS15145"
> table(x[, 3])
CDS exon start_codon stop_codon
794920 1309155 92839 83698
> y <- read.delim("~/igenomes/Homo_sapiens/NCBI/build37.2/Annotation/Genes/genes.gtf", header=FALSE, as.is=TRUE)
> table(y[, 3])
CDS exon start_codon stop_codon
345395 405671 34041 34012
> z <- read.delim("~/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf", header=FALSE, as.is=TRUE)
> table(z[, 3])
CDS exon start_codon stop_codon
365947 430178 36547 36532
> options(width=120)
> table(x[, 2])
3prime_overlapping_ncrna antisense IG_C_gene
92 40326 290
IG_C_pseudogene IG_D_gene IG_J_gene
40 196 78
IG_J_pseudogene IG_V_gene IG_V_pseudogene
12 1099 657
lincRNA miRNA misc_RNA
48588 6848 4380
Mt_rRNA Mt_tRNA nonsense_mediated_decay
4 44 293471
non_stop_decay polymorphic_pseudogene processed_pseudogene
1132 1853 24516
processed_transcript protein_coding pseudogene
172582 1978244 2247
retained_intron rRNA sense_intronic
150034 1140 2474
sense_overlapping snoRNA snRNA
1255 3242 4148
transcribed_processed_pseudogene transcribed_unprocessed_pseudogene translated_processed_pseudogene
1182 7931 3
TR_C_gene TR_D_gene TR_J_gene
58 9 246
TR_J_pseudogene TR_V_gene TR_V_pseudogene
8 869 107
unitary_pseudogene unprocessed_pseudogene
1467 13763
> table(y[, 2])
unknown
819119
> table(z[, 2])
unknown
869204
Another example. Read GTF file into R.
library(rtracklayer)
x <- import("/fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf")
x
# GRanges object with 1006819 ranges and 9 metadata columns:
length(unique(x$gene_name))
# [1] 26364
length(unique(x$transcript_id))
# [1] 54070
y <- read.delim("/fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf")
dim(y)
# [1] 1006818 9
colnames(y)
# [1] "chr1"
# [2] "unknown"
# [3] "exon"
# [4] "X11874"
# [5] "X12227"
# [6] "."
# [7] "X."
# [8] "..1"
# [9] "gene_id.DDX11L1..gene_name.DDX11L1..transcript_id.NR_046018..tss_id.TSS16932."
Convert transcript to symbol
- Check out XXX.genes.results or XXX.isoforms.results file from RSEM output
$ head Sample1.genes.results gene_id transcript_id(s) length effective_length expected_count TPM FPKM A1BG NM_130786 1766.00 1589.99 0.00 0.00 0.00 A1BG-AS1 NR_015380 2130.00 1953.99 0.00 0.00 0.00 $ head Sample1.isoforms.results transcript_id gene_id length effective_length expected_count TPM FPKM IsoPct NM_130786 A1BG 1766 1589.99 0.00 0.00 0.00 0.00 NR_015380 A1BG-AS1 2130 1953.99 0.00 0.00 0.00 0.00
- biotools (online)
- Converting between UCSC id and gene symbol with bioconductor annotation resources. You need to use the Homo.sapiens package.
library(Homo.sapiens) select(Homo.sapiens, "uc001qvk.1", "SYMBOL","TXNAME") # 'select()' returned 1:1 mapping between keys and columns # TXNAME SYMBOL # 1 uc001qvk.1 A2M
- How to map UCSC transcripts to gene symbol? org.Hs.eg.db package.
Splice junction
GTF file contains splice junction information. See Genome > Splice junction on how to extract this information.
bed
A BED file (.bed) is a tab-delimited text file that defines a feature track. Bed files can be like GTF/GFF to provide gene annotations. See ensembl.org. Tracks in the UCSC Genome Browser (http://genome.ucsc.edu/) can be downloaded to BED files and loaded into IGV.
A simple bed file looks like
chr7 127471196 127472363
The format is explained in UCSC.
We can also take a look some bed files generated after running the Tophat program.
brb@brb-T3500:~/Anders2013/Untreated-6$ head -n 3 insertions.bed track name=insertions description="TopHat insertions" 2L 10457 10457 T 1 2L 10524 10524 T 1 brb@brb-T3500:~/Anders2013/Untreated-6$ head -n 3 deletions.bed track name=deletions description="TopHat deletions" 2L 10832 10833 - 1 2L 12432 12434 - 1 brb@brb-T3500:~/Anders2013/Untreated-6$ head -n 3 junctions.bed track name=junctions description="TopHat junctions" 2L 11095 11479 JUNC00000001 4 - 11095 11479 255,0,0 2 74 ,70 0,314 2L 11271 11479 JUNC00000002 24 - 11271 11479 255,0,0 2 73 ,70 0,138
From my observations, the IGV can create the junctions track (it is called 'Untreated-6_s.bam junctions' when I load the <Untreated-6_s.bam> file) automatically once we load the bam file. If we manually load the <junctions.bed> file saved in the 'Untreated-6' directory, we can see both junction tracks are the same.
sam/bam, "samtools view" and Rsamtools
- http://biobits.org/samtools_primer.html
- Genome-SAMtools
- http://samtools.github.io/hts-specs/SAMv1.pdf (especially p4)
- http://genome.sph.umich.edu/wiki/SAM
- https://class.coursera.org/gencommand-003/lecture/39 (video)
- See VCF format
- https://youtu.be/2KqBSbkfhRo?list=UUqaMSQd_h-2EDGsU6WDiX0Q (Bioconductor)
There are 11 required fields in the sam file and the header is optional. See also http://bio-bwa.sourceforge.net/bwa.shtml.
| column field | example (/ExomeLungCancer/H1975cellLine) |
|---|---|
| 1 QNAME | SRR2923335.1 |
| 2 FLAG | 99 |
| 3 RNAME (chromosome name) | 6 |
| 4 POS | 36938157 |
| 5 MAPQ | 60 |
| 6 CIGAR | 100M |
| 7 RNEXT | = |
| 8 PNEXT | 36938259 |
| 9 TLEN | 202 |
| 10 SEQ | NGCAAGTAGAGGGGCGCACACCTACCCGCAGTTGTTCACAGCCATG… |
| 11 QUAL | #1=DDFDFHHHHHJIJJJJIJJJJJJJJIJJJJJIJIHHHHHFFF… |
| 12 Tags (optional) | NM:i:1 MD:Z:0G99 AS:i:99 XS:i:61 MQ:i:60 |
We can use samtools view to view the bam file (samtools view will not include the header, samtools view -H will include only the header, samtools view -h will include the header)
$ samtools view -h accepted_hits.bam | more @HD VN:1.4 @SQ SN:1 LN:249250621 @SQ SN:2 LN:243199373 @SQ SN:3 LN:198022430 @SQ SN:4 LN:191154276 @SQ SN:5 LN:180915260 @SQ SN:6 LN:171115067 @SQ SN:7 LN:159138663 @SQ SN:8 LN:146364022 @SQ SN:9 LN:141213431 @SQ SN:10 LN:135534747 @SQ SN:11 LN:135006516 @SQ SN:12 LN:133851895 @SQ SN:13 LN:115169878 @SQ SN:14 LN:107349540 @SQ SN:15 LN:102531392 @SQ SN:16 LN:90354753 @SQ SN:17 LN:81195210 @SQ SN:18 LN:78077248 @SQ SN:19 LN:59128983 @SQ SN:20 LN:63025520 @SQ SN:21 LN:48129895 @SQ SN:22 LN:51304566 @SQ SN:X LN:155270560 @SQ SN:Y LN:59373566 @SQ SN:MT LN:16569 @PG ID:STAR PN:STAR VN:STAR_2.5.0c CL:STAR --runThreadN 11 --genomeDir STARindex --readFilesIn test.SRR493369 _1.fastq test.SRR493369_2.fastq --outFileNamePrefix ./LFB_HOXA1KD_repA/ --outSAMtype BAM Unsorted --twop assMode Basic @CO user command line: STAR --genomeDir STARindex --twopassMode Basic --runThreadN 11 --outSAMtype BAM Unsorted --ou tFileNamePrefix ./LFB_HOXA1KD_repA/ --readFilesIn test.SRR493369_1.fastq test.SRR493369_2.fastq SRR493369.1 99 19 3977426 255 1S100M = 3977605 488 ACTGGATCTCCACAAGGTAGATGGGCTCCATGAGGCGTGG CTGGGCGGTCAGCACACTGGCGTAGAGGCAGCGCCGTGCTGTGGGGATGATCTGGCCCCCT CCCFFFFFHHHHHJJJJGIJJHIIJIJIJJIIJIJJJHJJJJJIIJDGDCDEDDDC DDCDDDB@BDDDDDDDBBB@99@BD>CCDD<@BCCCD:3@##### NH:i:1 HI:i:1 AS:i:199 nM:i:1 SRR493369.1 147 19 3977605 255 4M208N97M = 3977426 -488 CGCCCTCCTTGGTGGCCCACTGGAAGCCGGCC ACCACACTGTCCTTGATCTCGTTGAGGTACTGCACACCCTTGGTGATGTCGGTGAGGATGTTGGGGCCG 5BDDBBDDDDDDBABDDDCDDDDDDDCDDDDDCDCCCADEFFFFFHHH
To count the number of rows in the header, use the Linux comand grep (though samtools view provides the -H argument to keep only the header)
$ samtools view -h SeqTestdata/RNASeqFibroblast/LFB_HOXA1KD_repA/accepted_hits.bam | grep "^@" | wc -l $ # same as samtools view -H SeqTestdata/RNASeqFibroblast/LFB_HOXA1KD_repA/accepted_hits.bam | wc -l 28 $ samtools view -h SeqTestdata/RNASeqFibroblast/LFB_HOXA1KD_repA/accepted_hits.bam | grep -v "^@" | wc -l $ # same as samtools view SeqTestdata/RNASeqFibroblast/LFB_HOXA1KD_repA/accepted_hits.bam | wc -l 9856 $ samtools view -h SeqTestdata/RNASeqFibroblast/LFB_HOXA1KD_repA/accepted_hits.bam | wc -l 9884
We can also use Bioconductor's Rsamtools package to manipulate sam/bam, fasta, bcf and tabix files.
source("https://bioconductor.org/biocLite.R")
biocLite("Rsamtools")
library(Rsamtools)
bf1 = BamFileList(file = c("file1.bam", "file2.bam"))
seqinfo(bf1)
# Suppose we want to get the MAPQ value from reads
param<-ScanBamParam(what="mapq") # other choices, what = c("rname", "strand", "pos", "qwidth", "seq")
# see scanBamwhat()
bam<-scanBam(file="filename.bam", param=param)
str(bam)
summary(bam)
# List of 1
# $ :List of 1
# ..$ mapq: int [1:195875920] 0 13 0 0 2 0 0 0 9 0 ...
object.size(bam)/2^20
# 747.207664489746 bytes
summary(bam[[1]][[1]])
# Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
# 0.0 60.0 60.0 58.4 60.0 60.0 2434291
table(bam[[1]][[1]])
0 1 2 3 4 5 6 7
3186501 59642 40093 91541 52501 43220 123651 81155
8 9 10 11 12 13 14 15
23354 133441 19302 17924 61882 30326 17755 54093
16 17 18 19 20 21 22 23
24826 21290 52880 37874 15116 69845 25458 30183
24 25 26 27 28 29 30 31
57684 80430 13746 537584 18437 14125 41978 25748
32 33 34 35 36 37 38 39
11848 63802 13745 16987 33168 29840 12198 64799
40 41 42 43 44 45 46 47
763253 32690 61987 46020 64149 85350 129188 100040
48 49 50 51 52 53 54 55
193770 683051 64096 95453 58853 87942 58469 48584
56 57 58 59 60
50132 135261 70818 101295 185161256
Flag
https://broadinstitute.github.io/picard/explain-flags.html Interpret FLAG value (interactive)
For single end data, no flag is set so the flag is always 0.
Extract by read names with grep
# Extract reads SRR2923335.1 and SRR2923335.1999 and output only the read name (1st) and mapping quality columns (5th)
# -e = --extended-regexp, -w = matching the whole word
# awk print $1,$5 will output with a space character between columns
samtools view accepted_hits.bam | grep -E -w "SRR2923335.1|SRR2923335.1999" | awk '{print $1,$5}'
Extract by a region
# https://www.biostars.org/p/48719/ # the output will be all reads starting anywhere between 18000-45500. samtools view input.bam "Chr10:18000-45500" > output.bam # https://www.biostars.org/p/45936/ # index first!!! samtools view -h reads.bam 1:1042000-1042010 > subset.sam samtools view -h -b reads.bam chr1:10420000-10421000 > subset.bam
chromosomes
We can use the shell command in this post to extract the chromosome name, start position from the bam file. There we can see the difference of the chromosome name of bam files created from using Ensembl and UCSC.
# Ensembl GRCh37
brb@T3600 ~/SeqTestdata/RNASeqFibroblast $ samtools view LFB_scramble_repA/accepted_hits.bam | head | awk '{print $3}'
1
1
1
1
1
# NCBI GRCh38
brb@T3600 ~/SeqTestdata/RNASeqFibroblast $ samtools view RNAseq_Bam_ncbigrch38/LFB_scramble_repA.bam | head | awk '{print $3}'
chr1
chr1
chr1
chr1
chr1
chr1
# UCSC hg38
brb@T3600 ~/SeqTestdata/RNASeqFibroblast $ samtools view RNAseq_Bam_hg38/LFB_scramble_repA.bam | head | awk '{print $3}'
chr1
chr1
chr1
chr1
chr1
chr1
Subset reads from specific chromosomes
https://www.biostars.org/p/128967/
For example to remove mitochondrial reads from BAM files
samtools view -b input.bam chr1 chr2 chr3 chr4 > output.bam # Or if the bam file has been indexed (i.e. .bai file is available) samtools idxstats input.bam | cut -f 1 | grep -v MT | xargs samtools view -b input.bam > output.bam
where samtools idxstat returns 4 columns (TAB-delimited with each line consisting of reference sequence name, sequence length, # mapped reads and # unmapped reads).
$ samtools idxstats input.bam | head 1 249250621 0 0 2 243199373 0 0 3 198022430 0 0 4 191154276 0 0 5 180915260 0 0 6 171115067 0 0 7 159138663 0 0 8 146364022 0 0 9 141213431 0 0 10 135534747 0 0
Count the number of reads with certain number of mismatches
- https://www.biostars.org/p/130256/ Bwa appears to include only mismatches, but bowtie includes insertions and deletions in its NM.
- https://gist.github.com/davfre/8596159
Other alignment methods
Online tools (global and local alignments)
https://www.ebi.ac.uk/Tools/psa/
STAR
- Current Manual (pdf is embedded in github)
- Version 2.4 Manual (pdf is searchable)
Kallisto
Subread
See Subread.
Hisat
Command line usages
| Aligner | Command Line |
|---|---|
| bowtie2 | bowtie2 -x /path/to/bowtie2/BASENAME -1 A_1.fq,B_1.fq -2 A_2.fq,B_2.fq -S alignment.sam |
| tophat2 | tophat2 --no-coverage-search -p $SLURM_CPUS_PER_TASK \
-o OutputDir \ /path/to/bowtie2/BASENAME \ R1.FASTQ.gz R2.FASTQ.gz |
| STAR | mkdir STARindex
STAR --runMode genomeGenerate \ --runThreadsN $SLURM_CPUS_PER_TASK \ --genomeDir STARindex \ --genomeFastaFiles /path/to/fasta.fa \ --sjdbGTFfile /path/to/genes.gtf \ --sdjbOverhang 85 STAR --twopassMode Basic \ --runThreadN $SLURM_CPUS_PER_TASK \ --genomeDir STARindex \ --outSAMmapqUnique 50 \ --sjdbOverhang 85 \ --readFilesIn r1.fastq.gz r2.fastq.gz \ --readFilesCommand zcat \ --outSAMtype BAM Unsorted \ --outFileNamePrefix output/star |
| Subread | subread-buildindex -o INDEX /path/to/fasta.fa
subread-align -T $SLURM_CPUS_PER_TASK -i INDEX -r R1.FASTQ.gz -R R2.FASTQ.gz -o output/alignment.sam |
| Hisat2 | hisat2-build -p $SLURM_CPUS_PER_TASK \
/path/to/fasta.fa INDEX hisat2 -p $SLURM_CPUS_PER_TASK -x INDEX \ -1 R1.FASTQ.gz \ -2 R2.FASTQ.gz -S output/alignment.sam |
What's special with rna-seq count data
Overdispersion
- http://genomicsclass.github.io/book/pages/rnaseq_gene_level.html
- Dispersion Estimation and Its Effect on Test Performance in RNA-seq Data Analysis: A Simulation-Based Comparison of Methods by Landau 2013.
- Gene dispersion is the key determinant of the read count bias in differential expression analysis of RNA-seq data
Normalization
- The Impact of Normalization Methods on RNA-Seq Data Analysis by Zyprych-Walczak 2015. Two public datasets (Bodymap and Cheung) are available from recount website.
- Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments by Bullard 2010.
- Comparing the normalization methods for the differential analysis of Illumina high-throughput RNA-Seq data by Peipei Li 2015.
- Comparison of TMM (edgeR), RLE (DESeq2), and MRN Normalization Methods by Maza E 2016
- A Hypothesis Testing Based Method for Normalization and Differential Expression Analysis of RNA-Seq Data by Zhou 2017
Power Calculation
- Power analysis and sample size estimation for RNA-Seq differential expression Ching 2014
- PROPER: comprehensive power evaluation for differential expression using RNA-seq Wu 2015
- Extending the R Library PROPER to Enable Power Calculations for Isoform-Level Analysis with EBSeq Gaye 2016
- Power analysis for RNA-Seq differential expression studies Yu 2017
Phenotype Prediction
Improving the value of public RNA-seq expression data by phenotype prediction
Clustering
ICAclust – Independent Component Analysis (ICA) based-clustering of temporal RNA-seq data
Correlation between RNA-Seq and microarrays
Correlation between RNA-Seq and microarrays results using TCGA data
Techniques for characterizing RNA transcripts
qRT-PCR -> Microarray -> Nanostring -> RNA-Seq
Integrating RNA sequencing into neuro-oncology practice
Differential expression analysis
- How not to perform a differential expression analysis (or science)
- A Comparative Study of Techniques for Differential Expression Analysis on RNA-Seq Data (Zhang, 2014)
Reproducible RNA-Seq analysis
Other RNA-Seq data
GEO
Go to http://www.ncbi.nlm.nih.gov/sites/entrez and select 'GEO Datasets' from the drop down menu, and use 'RNA-Seq' as your search term. http://www.ncbi.nlm.nih.gov/gds?term=rna-seq
GSE28666
as used in DAFS paper. The timing is
- bowtie2-build 115 minutes
- fastq-dump 25 minutes per fastq (3.3 - 5.9GB fastq files)
- tophat2 90 minutes per fastq
- samtools 5 minutes
- htseq-count 13 minutes
GSE30567
Used in STAR paper
GSE38886
Used in STAR paper
GSE48215 & GSE48216
Modeling precision treatment of breast cancer
GSE48215 is Whole exome Seq and GSE48216 is RNA-Seq
GSE51403 (full R script)
RNA-seq differential expression studies: more sequence, or more replication?
An example of R code to download SRA files.
homo sapiens,
Illumina HiSeq 2000.
Question: what reference genome I should use?
Ans1: hg18 was used.
See http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSM1244821
Or the paper.
Ans2: GRCh38:
Click 'BioProject' in http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51403
then click 'Genome' in http://www.ncbi.nlm.nih.gov/bioproject/PRJNA222975
But tophat website only provides GRCh37.
https://genome.ucsc.edu/goldenPath/newsarch.html shows GRCh38 assembly is released
in Dec/2013.
38 fastq/SRR (all are single lane)
14 samples/GSM
GSM# SRX# Sample SRR#
---------------------------------------------------------------
Ctrl 1244822 365217 Ctrl_Rep7 1012952, 1012953, 1012954 (3 runs, 70M spots, 3.5G bases)
1244821 365216 Ctrl_Rep6 949-951 (I just skip prefix '1012')
1244820 365215 Ctrl_Rep5 946-948 (3 runs)
1244819 365214 Ctrl_Rep4 943-945 (3 runs) Control ethanol 24h
1244818 365213 Ctrl_Rep3 940-942 (3 runs)
1244817 365212 Ctrl_Rep2 937-939 (3 runs)
1244816 365211 Ctrl_Rep1 934-936 (3 runs) Control ethanol 24h
Trmnt 1244815 365210 E2_Rep7 931-933 (3 runs) 10nM E2 treatment for 24h
1244814 365209 E2_Rep6 928-930 (3 runs)
1244813 365208 E2_Rep5 925-927 (3 runs)
1244812 365207 E2_Rep4 923-924 (2 runs)
1244811 365206 E2_Rep3 921-922 (2 runs)
1244810 365205 E2_Rep2 919-920 (2 runs) 10nM E2 treatment for 24h
1244809 365204 E2_Rep1 917-918 (2 runs) 10nM E2 treatment for 24h
# Step 1. Download and convert data
library(SRAdb)
setwd("~/GSE51403")
if( ! file.exists('SRAmetadb.sqlite') ) {
# sqlfile <- getSRAdbFile()
sqlfile <- "~/Anders2013/SRAmetadb.sqlite"
} else {
sqlfile <- 'SRAmetadb.sqlite'
}
sra_con <- dbConnect(SQLite(),sqlfile)
fs <- listSRAfile("SRP031476", sra_con, fileType = "sra" ) # 38 files
# getSRAfile("SRP031476", sra_con, fileType = "sra" ) # starting to download
getSRAfile(fs$run[13], sra_con, fileType='sra') # SRR1012917.sra
getSRAfile(fs$run[30], sra_con, fileType='sra') # SRR1012918.sra
dbDisconnect( sra_con )
fs <- dir(pattern='sra')
# for(f in fs) system(paste("fastq-dump --split-3", f)) # Single thread, Not efficient
scrp <- paste("/opt/RNA-Seq/bin/sratoolkit.2.3.5-2-ubuntu64/bin/fastq-dump --split-3", fs)
library(parallel)
cl <- makeCluster(getOption("cl.cores", 5))
# note that if something is wrong, we need to delete broken fastq first and then run again.
system.time(clusterApply(cl, scrp, function(x) system(x)))
stopCluster(cl)
# Step 2. Create samples object
samples <- data.frame(LibraryName = c(paste("Ctrl_Rep", 7:1, sep=''),
paste("E2_Rep", 7:1, sep='')),
LibraryLayout = rep("SINGLE", 14),
fastq1=c(paste(paste("SRR", 1012952:1012954, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012949:1012951, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012946:1012948, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012943:1012945, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012940:1012942, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012937:1012939, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012934:1012936, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012931:1012933, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012928:1012930, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012925:1012927, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012923:1012924, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012921:1012922, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012919:1012920, ".fastq", sep=''), collapse=','),
paste(paste("SRR", 1012917:1012918, ".fastq", sep=''), collapse=',')),
fastq2=rep("", 14),
countf=c(paste("Ctrl_Rep", 7:1, ".count", sep=''),
paste("E2_Rep", 7:1, ".count", sep='')))
# Step 3: run tophat2
i <- 10
system.time(paste("tophat2 -G genes.gtf -p 5 -o",
samples$LibraryName[i],
"genome",
samples$fastq1[i]))
# system("tophat2 -G genes.gtf -p 5 -o E2_Rep5 genome SRR1012925.fastq")
# system("tophat2 -G genes.gtf -p 5 -o E2_Rep5 genome SRR1012925.fastq,SRR1012926.fastq,SRR1012927.fastq")
for(i in c(1:14)) {
system(paste("tophat2 -G genes.gtf -p 5 -o", samples$LibraryName[i], "genome", samples$fastq1[i]))
}
# Step 4. Organize, sort and index the BAM files and create SAM files.
for(i in seq_len(nrow(samples)) {
lib = samples$LibraryName[i]
ob = file.path(lib, "accepted_hits.bam")
# sort by name, convert to SAM for htseq-count
c1 <- paste0("samtools sort -n ", ob, " ", lib, "_sn")
system(c1)
}
samtools sort -n Sample1/accepted_hits.bam Sample1_sn # 283 seconds
# samtools sort Sample1/accepted_hits.bam Sample1_s
# Step 5. Count reads using htseq-count
htseq-count -f bam -s no -a 10 Sample1_sn.bam Mus_musculus.GRCm38.70.gtf > Sample1.count # 759 seconds
htseq-count -f bam -s no -a 10 Sample2_sn.bam Mus_musculus.GRCm38.70.gtf > Sample2.count
samples$countsf <- paste(samples$LibraryName, "count", sep=".")
gf <- "genes.gtf"
cmd <- paste0("htseq-count -s no -a 10 ", samples$LibraryName, "_sn.sam ", gf, " > ", samples$countf)
system.time(clusterApply(cl, cmd, function(x) system(x))) # 119 minutes
samplesDESeq = data.frame(shortname = c("Sample1", "Sample2"), countf=c("Sample1.count", "Sample2.count"),
condition = c("CTL", "KD"), LibraryLayout =c("SINGLE", "SINGLE"))
library("DESeq")
cds = newCountDataSetFromHTSeqCount(samplesDESeq)
cds = estimateSizeFactors(cds)
sizeFactors(cds)
GSE37544
GSE11045
Probe Region Expression Estimation for RNA-Seq Data for Improved Microarray Comparability
GSE37703/SRP012607
Differential analysis of HOXA1 in adult cells at isoform resolution by RNA-Seq by Trapnell C, Hendrickson DG, Sauvageau M, Goff L et al. Nat Biotechnol 2013.
The <samples.txt> file required by BRB-SeqTools looks like
LibraryName LibraryLayout fastq1 fastq2 ReadGroup SequencerManufacturer LFB_scramble_repA PAIRED SRR493366_1.fastq SRR493366_2.fastq 1 illumina LFB_scramble_repB PAIRED SRR493367_1.fastq SRR493367_2.fastq 2 illumina LFB_scramble_repC PAIRED SRR493368_1.fastq SRR493368_2.fastq 3 illumina LFB_HOXA1KD_repA PAIRED SRR493369_1.fastq SRR493369_2.fastq 4 illumina LFB_HOXA1KD_repB PAIRED SRR493370_1.fastq SRR493370_2.fastq 5 illumina LFB_HOXA1KD_repC PAIRED SRR493371_1.fastq SRR493371_2.fastq 6 illumina
GSE37704
http://www.gettinggeneticsdone.com/2015/12/tutorial-rna-seq-differential.html
GSE64570, GSE69244, GSE72165
Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences
Ion Torrent
- GEO
- GSE46876 from GPL17301 PGM (Homo sapiens) and GPL17303 Ion Torrent Proton (Homo sapiens). Unequal read length.
- GSE83668 from GPL17303 Ion Torrent Proton (Homo sapiens) and GPL11154 Illumina HiSeq 2000 (Homo sapiens)
- GSE79051/SRP071553 from GPL17303 Ion Torrent Proton (unequal read length)
SRA
Pipeline
Extracting allelic read counts from 250,000 human sequencing runs in Sequence Read Archive 2018. Code in Github and Data.
SRA000299
Note that one fastq file can represent several channels/lanes. The lane information can be extracted from fastq definition line (first line of each read). For example,
@SRR002321.1 080226_CMLIVERKIDNEY_0007:2:1:115:885
"SRR002321.1" is the short read archive name, where the .1 is the read number, "080226_CMLIVERKIDNEY_0007" should be the Machine name, which has been arbitrarily changed
- "2" is the Channel/lane number
- "1" is the Tile number
- "115" is the X position
- "885" is the Y position
The website also shows how to extract reads with the same run from fastq files.
In this example, there are
- two full sequencing runs (this info can be obtained from paper)
- 4 experiments (SRX000571, SRX000604, SRX000605, SRX000606)
- 6 runs (SRRR002321, SRR002323, SRR002322, SRR002320, SRR002325, SRR002324)
- 7 lanes (eg "080226_CMLIVERKIDNEY_0007:2") for each full sequencing run
SRA data linked from QuickNGS
Include Chip-Seq, RNA-Seq, WGS and miRNA-Seq. QuickNGS is a production environment for quick batch-wise analysis of Next-Generation Sequencing (NGS) data in high-throughput laboratories.
European Nucleotide Archive/ENA
DDBJ
ENCODE
http://www.ncbi.nlm.nih.gov/geo/info/ENCODE.html has a list of data with GSE number.
1000 genomes
Human BodyMap 2.0 data from Illumina
- http://www.ensembl.info/blog/2011/05/24/human-bodymap-2-0-data-from-illumina/
- https://usegalaxy.org/u/jeremy/p/galaxy-rna-seq-analysis-exercise. Galaxy needs to install hg19 genome before we can use tophat or bowtie. See Galaxy/Wiki/Admin/Data Preparation page. Genome can be downloaded from tophat webpage. It is 20GB for hg19! We can also download just chromosome 19 fasta file chr19.fa.gz from UCSC genomes FTP server and it is only 19MB. We can then create reference index files (.bt2) by using bowtie2-build command. If we use Galaxy, we don't need to create reference index files.
- Impact of gene annotation on RNA-Seq data analysis
TCGA/The Cancer Genome Atlas
- Broad Institute TCGA GDAC
- https://wiki.nci.nih.gov/display/TCGA/RNASeq
- https://www.biostars.org/p/128132/
- TCGAretriever from CRAN
- RTCGA package from Bioconductor.
- RTCGA.clinical - Clinical datasets from The Cancer Genome Atlas Project
- Extract Survival Information from Datasets Included in RTCGA.clinical and RTCGA.clinical.20160128 Packages
- https://rtcga.github.io/RTCGA/reference/expressionsTCGA.html
> if (!requireNamespace("BiocManager", quietly = TRUE))
install.packages("BiocManager")
> BiocManager::install("RTCGA.clinical")
> library(RTCGA.clinical)
# Available cohorts
> checkTCGA('Dates')
[1] "2011-10-26" "2011-11-15" "2011-11-28" "2011-12-06" "2011-12-30"
[6] "2012-01-10" "2012-01-24" "2012-02-17" "2012-03-06" "2012-03-21"
[11] "2012-04-12" "2012-04-25" "2012-05-15" "2012-05-25" "2012-06-06"
[16] "2012-06-23" "2012-07-07" "2012-07-25" "2012-08-04" "2012-08-25"
[21] "2012-09-13" "2012-10-04" "2012-10-18" "2012-10-20" "2012-10-24"
[26] "2012-11-02" "2012-11-14" "2012-12-06" "2012-12-21" "2013-01-16"
[31] "2013-02-03" "2013-02-22" "2013-03-09" "2013-03-26" "2013-04-06"
[36] "2013-04-21" "2013-05-08" "2013-05-23" "2013-06-06" "2013-06-23"
[41] "2013-07-15" "2013-08-09" "2013-09-23" "2013-10-10" "2013-11-14"
[46] "2013-12-10" "2014-01-15" "2014-02-15" "2014-03-16" "2014-04-16"
[51] "2014-05-18" "2014-06-14" "2014-07-15" "2014-09-02" "2014-10-17"
[56] "2014-12-06" "2015-02-02" "2015-02-04" "2015-04-02" "2015-06-01"
[61] "2015-08-21" "2015-11-01" "2016-01-28"
# Available cohorts
> (cohorts <- infoTCGA() %>% rownames() %>% sub("-counts", "", x=.))
[1] "ACC" "BLCA" "BRCA" "CESC" "CHOL" "COAD"
[7] "COADREAD" "DLBC" "ESCA" "FPPP" "GBM" "GBMLGG"
[13] "HNSC" "KICH" "KIPAN" "KIRC" "KIRP" "LAML"
[19] "LGG" "LIHC" "LUAD" "LUSC" "MESO" "OV"
[25] "PAAD" "PCPG" "PRAD" "READ" "SARC" "SKCM"
[31] "STAD" "STES" "TGCT" "THCA" "THYM" "UCEC"
[37] "UCS" "UVM"
> dim(BRCA.clinical)
[1] 1098 3703
> dim(OV.clinical) # one row per sample
[1] 591 3626
> dim(LUAD.clinical)
[1] 522 3046
> OV.survInfo <- survivalTCGA(OV.clinical, extract.cols = "admin.disease_code")
> dim(OV.survInfo) # 576 x 4
> plot(survfit(Surv(times, patient.vital_status) ~ 1,
data = OV.survInfo), conf.int=F, mark.time=TRUE)
> BiocManager::install("RTCGA.mRNA")
> data(package = "RTCGA.mRNA")
> data(OV.mRNA, package = "RTCGA.mRNA")
> dim(OV.mRNA) # one row per sample
[1] 561 17815
> OV.mRNA[1:2, 1:5]
bcr_patient_barcode ELMO2 CREB3L1 RPS11 PNMA1
1 TCGA-01-0628-11A-01R-0363-07 -0.24800000 -0.14150 1.370625 1.08775
2 TCGA-01-0639-11A-01R-0363-07 0.09508333 -0.04650 0.982875 1.38725
> OV.clinical[1:2, 18]
[1] "tcga-04-1331" "tcga-04-1332"
> match(substr(OV.mRNA[, 1], 6, 12), substr(OV.clinical[, 18], 6, 12)) %>%
na.omit() %>% length()
[1] 546
The TCGA Data Portal does not host lower levels of sequence data. NCI's Cancer Genomics Hub (CGHub) is the new secure repository for storing, cataloging, and accessing BAM files and metadata for sequencing data (need to 1. register from HHS to get an access 2. request access to the secure data set).
UCI
- Machine learning repository (not RNA-Seq)
ReCount
- ReCount: A multi-experiment resource of analysis-ready RNA-seq gene count datasets
- http://bowtie-bio.sourceforge.net/recount/
- Two datasets were used in Getting the most out of RNA-seq data analysis
Other RNA-Seq tools
http://en.wikipedia.org/wiki/List_of_RNA-Seq_bioinformatics_tools
Galaxy
The first 3 lines will install galaxy on local computer. The last command will start Galaxy. PS. It will take a while to start the first time.
hg clone https://bitbucket.org/galaxy/galaxy-dist/ cd galaxy-dist hg update stable sh run.sh
A screenshot of the available tools is given below. Strangely, I did not find "NGS: RNA Analysis" in the NGS: ToolBox on left hand side.
We cannot upload > 2GB fastq data. For such large dataset (we have one 3.3 GB fastq in this case), Galaxy asks to use http or ftp to load the data. I install Apache in Ubuntu and put this fastq file there for upload.
Galaxy does not include bowtie, tophat, .... by default. See this post and Galaxy wiki about the installation. Another option is to modify line 614 of 'universe_wsgl.ini' file and enable ourselves as adminstrator. Also modify line 146 to add a path for installing tools through Galaxy Admin page.
If you submit a lot of jobs (eg each tophat2 generates 5 jobs) some of jobs cannot be run immediately. In my experience, there is a 25 jobs cap. Running jobs has a yellow background color. Waiting for run jobs has a gray background color.
Some helpful information
- Virtual Appliances
- Galaxy Docker Image
- Quick & dirty Galaxy installation in a virtual machine
- Downloadable Galaxy Virtual Machine In Vmware. The virtual machine is not there anymore but the documentation is still there.
- https://wiki.galaxyproject.org/CitingGalaxy Citation
- https://wiki.galaxyproject.org/Learn Search rna-seq or NGS as keywords.
- https://biostar.usegalaxy.org/
- Galaxy & DESeq2
- Galaxy Demo -- RNAseq Pipeline as Example from BioIT Core.
Cufflink -> Cuffcompare or Cuffmerge -> Cuffdiff
- http://cole-trapnell-lab.github.io/cufflinks/
- UCSF wiki.
- Michigan State University
- http://seqanswers.com/forums/showthread.php?t=9753
- https://docs.uabgrid.uab.edu/wiki/UAB_Galaxy_RNA_Seq_Step_by_Step_Tutorial#Cufflinks_inputs
- https://sites.google.com/site/princetonhtseq/tutorials/rna-seq
- http://www.genomecenter.ucdavis.edu/resources/instructional-videos
Forum
recount2
A multi-experiment resource of analysis-ready RNA-seq gene and exon count datasets https://jhubiostatistics.shinyapps.io/recount/
recount3
RNACocktail
A comprehensive framework for accurate and efficient RNA-Seq analysis
Massive Mining of Publicly Available RNA-seq Data from Human and Mouse
- ARCHS4: All RNA and ChIP-Seq Sample and Signature Search
- ARCHS4 tutorial (youtube). The chrome extension did not work on GSE26880 (still no ARCHS4 icon on the GEO webpage after restart Chrome. Click the series matrix file link, it brings to a page to download 3 GPL series matrix. But the series matrix files still don't contain the expression)
Misc
Markets
Global And United States NGS-based RNA-seq Market 2017
Tutorial/Primer
Whole Transcriptome Profiling: An RNA-Seq Primer and Implications for Pharmacogenomics Research