R parallel
Introduction
- Example code for the book Parallel R by McCallum and Weston.
- A gentle introduction to parallel computing in R
- An introduction to distributed memory parallelism in R and C
- Processing: When does it worth?
- Parallel Computing With R: A Brief Review by Dirk Eddelbuettel (updated 2020).
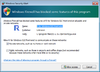
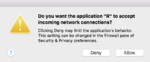
Security warning from Windows/Mac
It seems it is safe to choose 'Cancel' when Windows Firewall tried to block R program when we use makeCluster() to create a socket cluster.
library(parallel)
cl <- makeCluster(2)
clusterApply(cl, 1:2, get("+"), 3)
stopCluster(cl)
If we like to see current firewall settings, just click Windows Start button, search 'Firewall' and choose 'Windows Firewall with Advanced Security'. In the 'Inbound Rules', we can see what programs (like, R for Windows GUI front-end, or Rserve) are among the rules. These rules are called 'private' in the 'Profile' column. Note that each of them may appear twice because one is 'TCP' protocol and the other one has a 'UDP' protocol.
Exit with care when running parallel within function
If the parallel code is within function, it is possible that function ends as the result of an error and so stopCluster() is omitted. The solution is to use on.exit() function. See Efficient R programming → Parallel computing.
foo <- function(cores) {
cl <- makeCluster(cores)
on.exit(stopCluster(cl))
# Do something
}
Detect number of cores and memory consideration
parallel::detectCores() # not good future::availableCores() # good
Don't use the default option getOption("mc.cores", 2L) (PS it only returns 2.) in mclapply() unless you are a developer for a package.
However, it is a different story when we run the R code in HPC cluster. Read the discussion Whether to use the detectCores function in R to specify the number of cores for parallel processing?
On NIH's biowulf, even I specify an interactive session with 4 cores, the parallel::detectCores() function returns 56. This number is the same as the output from the bash command grep processor /proc/cpuinfo or (better) lscpu. The free -hm also returns a full 125GB size instead of my requested size (4GB by default). The future::availableCores() fixes the problem.
See Biowulf's R webpage for a detailed instruction and my note Running R scripts for selecting the number of cores on biowulf/non-biowulf (different OS).
parallelly 1.31.1: Better at Inferring Number of CPU Cores with Cgroups and Linux Containers. Apr 2022
Please Avoid detectCores() in your R Packages Dec 2022
Identify OS
See Distinguish Windows, Linux, Mac
Memory
- Alternative to R's `memory.size()` in linux? pryr::mem_used() will return the memory currently used by R. pryr::address() to get the address of the variable in the cluster.
- Use makeCluster(no_cores, type = "FORK") instead of makeCluster(no_cores, type = "SOCK"). See How-to go parallel in R – basics + tips.
- system('grep MemTotal /proc/meminfo', intern = TRUE) will show the total physical RAM on Linux OS (mac does not work).
- Monitor memory usage with R parallel. profvis package from RStudio.
- It works on RStudio or a regular R session
- The output is an HTML file
- We can change the interval period. Its default is 0.01 seconds or 100 ms. profvis(expr = NULL, interval = 0.01, ...)
- When I test profvis() on a function which calls clusterExport() and parSapply(), I can see the time spent on each of them. My example of running coxph on a set of genes.
- Question: why the sum of time from the HTML output not close at all to system.time() when parLapply() is used?
- Running time under-estimated with parallel::parLapply(). The profiler only considers the main thread. Which means that any thread/process spawned from the main rsession process, will not be accounted for in the execution time. Only the thread that spawns the workers are timed
- Profile non-cpu time?
- Advanced R > Measuring performance
- Efficient R programming > Code profiling.
- Rprof() and summaryRprof()
Rprof(tf <- "rprof.txt", memory.profiling = TRUE) # the code you want to profile must be in between Rprof (NULL) ; print(summaryRprof(tf))
Socket or Fork
- For the fork, each parallel thread is a complete duplication of the master process with the shared environment, including objects or variables defined prior to the kickoff of parallel threads. Therefore, it runs fast. However, the major limitation is that the fork doesn’t work on the Windows system.
- The socket works on all operating systems. Each thread runs separately without sharing objects or variables, which can only be passed from the master process explicitly. As a result, it runs slower due to the communication overhead.
install.packages(c("rbenchmark", "nycflights13"))
data(flights, package = 'nycflights13')
dim(flights) # [1] 336776 19
ex <- expression(carrier == "UA" & origin == "EWR" & day == 1 & is.na(arr_time))
parFilter <- function(df, ex, type) {
cn <- parallel::detectCores() - 1
cl <- parallel::makeCluster(cn, type = type)
### DIVIDE THE DATAFRAME BASED ON # OF CORES
sp <- parallel::parLapply(cl, parallel::clusterSplit(cl, seq(nrow(df))),
function(c_) df[c_,])
### PASS THE OBJECT FROM MASTER PROCESS TO EACH NODE
parallel::clusterExport(cl, "ex")
### EXTRACT ROW INDEX ON EACH NODE
id <- Reduce(c, parallel::parLapply(cl, sp,
function(s_) with(s_, eval(ex))))
parallel::stopCluster(cl)
return(df[which(id),])
}
rbenchmark::benchmark(replications = 10, order = "elapsed", relative = "elapsed",
columns = c("test", "replications", "elapsed", "relative"),
" FORK" = parFilter(flights, ex, "FORK"),
"SOCKET" = parFilter(flights, ex, "PSOCK")
) # Phenom(tm) II X6 1055T
test replications elapsed relative
1 FORK 10 42.154 1.000
2 SOCKET 10 68.901 1.635
makeCluster() for socket connection in R 4.0
The function hangs on R 4.0.0 + RStudio. See
- https://github.com/rstudio/rstudio/issues/6692 for a simple solution cl <- makeCluster(ncores, setup_strategy = "sequential")
- The article Socket Connections Update for R 4.0
How can I print when using %dopar%
https://stackoverflow.com/a/15078540
Create and register your cluster with something like:
library(doParallel)
cl<-makeCluster(4, outfile = "debugParallel.txt")
registerDoParallel(cl)
foreach(x=list(1, 2, "a")) %dopar%
{
print(x) # Sys.getpid() # print process ID
}
stopCluster(cl)
And the debugParallel.txt will look like
starting worker pid=17718 on localhost:11704 at 15:10:29.665 starting worker pid=17733 on localhost:11704 at 15:10:29.850 starting worker pid=17747 on localhost:11704 at 15:10:30.025 starting worker pid=17761 on localhost:11704 at 15:10:30.183 [1] 1 [1] 2 [1] "a"
Setting outfile to the empty string ("") will prevent doParallel (or snow whatever) from redirecting the output, often resulting in the output from your print messages showing up on the terminal of the master process. Your foreach loop doesn't need to change at all.
parallel package (including parLapply, parSapply)
Parallel package was included in R 2.14.0. It is derived from the snow and multicore packages and provides many of the same functions as those packages.
The parallel package provides several *apply functions for R users to quickly modify their code using parallel computing.
- makeCluster(makePSOCKcluster, makeForkCluster), stopCluster. Other cluster types are passed to package snow.
- clusterCall, clusterEvalQ: source R files and/or load libraries
- clusterSplit
- clusterApply, clusterApplyLB (vs the foreach package)
- clusterExport: export variables. We may need to apply the option envir=environment() if the objects to be exported are local variables.
- clusterMap
- parallel::mclapply() and parallel::parLapply()
- parLapply, parSapply, parApply, parRapply, parCapply. Note that
- parSapply() can be used to as a parallel version of the replicate() function. See this example or this example.
- An iteration parameter needs to be added to the first parameter of the main function.
- parLapplyLB, parSapplyLB (load balance version)
- clusterSetRNGStream, nextRNGStream, nextRNGSubStream
Examples (See ?clusterApply)
library(parallel)
cl <- makeCluster(2, type = "SOCK")
clusterApply(cl, 1:2, function(x) x*3) # OR clusterApply(cl, 1:2, get("*"), 3)
# [[1]]
# [1] 3
#
# [[2]]
# [1] 6
parSapply(cl, 1:20, get("+"), 3)
# [1] 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23
stopCluster(cl)
An example of using clusterCall() or clusterEvalQ()
library(parallel)
cl <- makeCluster(4)
clusterCall(cl, function() {
source("test.R")
})
# clusterEvalQ(cl, {
# source("test.R")
# })
## do some parallel work
stopCluster(cl)
mclapply() (Windows OS only supports 1 core, forking) vs parLapply()/parSapply() (all OSs, clustering)
- Be providing the number of cores in mclapply() using mc.cores argument (2 is used by default)
- set.seed() can not fix seed in mclapply(); we need to use set.seed(, "L'Ecuyer-CMRG"). But be careful on the side-effect of using L'Ecuyer-CMRG seed.
- Chapter 4 of Parallel R by Stephen Weston, Q. Ethan McCallum
- Setting a seed in R, when using parallel simulation
- R doesn't reset the seed when “L'Ecuyer-CMRG” RNG is used?
library(parallel) system.time(mclapply(1:1e4L, function(x) rnorm(x))) system.time(mclapply(1:1e4L, function(x) rnorm(x), mc.cores = 4)) set.seed(1234) mclapply(1:3, function(x) rnorm(x)) set.seed(1234) mclapply(1:3, function(x) rnorm(x)) # cannot reproduce the result mclapply(1:3, function(x) {set.seed(x); rnorm(x)}) # Can reproduce the result set.seed(123, "L'Ecuyer") mclapply(1:3, function(x) rnorm(x)) mclapply(1:3, function(x) rnorm(x)) # results are not changed once we have run set.seed( , "L'Ecuyer") set.seed(1234) # use set.seed() in order to get a new reproducible result mclapply(1:3, function(x) rnorm(x)) mclapply(1:3, function(x) rnorm(x)) # results are not changed # an example from ?mclapply library(boot) cd4.rg <- function(data, mle) MASS::mvrnorm(nrow(data), mle$m, mle$v) cd4.mle <- list(m = colMeans(cd4), v = var(cd4)) mc <- getOption("mc.cores", 2) run1 <- function(...) boot(cd4, corr, R = 500, sim = "parametric", ran.gen = cd4.rg, mle = cd4.mle) ## To make this reproducible: set.seed(123, "L'Ecuyer") res <- mclapply(seq_len(mc), run1) cd4.boot <- do.call(c, res) boot.ci(cd4.boot, type = c("norm", "basic", "perc"), conf = 0.9, h = atanh, hinv = tanh) - An example of applying to the permutation test (LS test for GSEA)
ngenes <- 10000 nresamp <- 1e5 genesetSize <- 20 pinter <- runif(ngenes) lsobs <- -mean(log(pinter[genesetSize])) system.time( lsperm <- mclapply(1:nresamp, function(i) -mean(log(sample(pinter, genesetSize)))) ) # system.time( lsperm <- mclapply(1:nresamp, function(i) -mean(log(sample(pinter, genesetSize))), mc.cores = 4) ) lsperm <- unlist(lsperm) mean(lsperm > lsobs)
- tempdir() is per session only. So the following will give the same directory.
parallel::mclapply(1:3, function(x) tempdir())
- An example of using parallel::mcMap().
Note
- Windows OS can not use mclapply(). The mclapply() implementation relies on forking and Windows does not support forking. mclapply from the parallel package is implemented as a serial function on Windows systems. The parallelsugar package was created based on the above idea.
- Another choice for Windows OS is to use parLapply() function in parallel package.
- Understanding the differences between mclapply and parLapply in R You don't have to worry about reproducing your environment on each of the cluster workers if mclapply() is used.
ncores <- as.integer( Sys.getenv('NUMBER_OF_PROCESSORS') ) cl <- makeCluster(getOption("cl.cores", ncores)) LLID2GOIDs2 <- parLapply(cl, rLLID, function(x) { library(org.Hs.eg.db); get("org.Hs.egGO")[[x]]} ) stopCluster(cl)It does work. Cut the computing time from 100 sec to 29 sec on 4 cores.
mclapply Errors
scheduled core 1 did not deliver a result, all values of the job will be affected
mclapply() vs parLapply() vs foreach()
- One of my examples shows mclapply() used much more memory than other methods
- mclapply() uses 10.5GB while parLapply() uses around 6.5GB and foreach() (either doParallel or doSNOW backend) uses about 7GB (the numbers are obtained by monitoring the output from jobload command). NB: the memory used from the output of jobhist command in Biowulf is not reliable
- They are taking about the same amount of running time (19 minutes)
- mclapply() uses fork. So it does not need clusterExport() nor clusterEvalQ() while other methods like parLapply() need.
- Bottomline: Do not use mclapply() if we want to save memory
- The code is available in my private bitbucket repository
- https://stackoverflow.com/questions/44806048/r-mclapply-vs-foreach
- mclapply does NOT copy the complete workspace contents to the child processes
- Trying to reduce the memory overhead when using mclapply. mclapply is the simplest (no need to export objects nor load packages) but it has a very high memory load.
- Parallel programming in R by Bjørn-Helge Mevik. parLapply() works on MPI for multiple machines.
| for loop | parallel::parLapply |
|---|---|
out <- vector("list", 10)
onerun <- function(j) { set.seed(seeds[j]) }
for(j in 1:10) {
out[[j]] <- onerun(j)
}
|
require(parallel)
ncpus <- 4
cl <- makeCluster(ncpus)
clusterEvalQ(cl, {
library(glmnet)
source("foo.R")
})
clusterExport(cl, c("seeds"))
onerun <- function(j) { set.seed(seeds[j]) }
out <- parLapply(cl, 1:10, onerun)
stopCluster(cl)
|
clusterApply() vs parLapply() and load-balancing versions
Look at the documentation clusterApply, it seems there is no difference in these two.
Note that there are load-balancing versions of these two: clusterApplyLB() and parLapplyLB().
Why does parLapplyLB not actually balance load? (Now only relevant for R versions < 3.5.0)
parallel vs doParallel package
doParallel is a foreach parallel adaptor for the parallel package.
doParallel provides a parallel backend for the %dopar% function using the parallel package.
doParallel depends on foreach, iterators, parallel and utils packages.
parallelsugar package
If we load parallelsugar, the default implementation of parallel::mclapply, which used fork based clusters, will be overwritten by parallelsugar::mclapply, which is implemented with socket clusters.
library(parallel)
system.time( mclapply(1:4, function(xx){ Sys.sleep(10) }) )
## user system elapsed
## 0.00 0.00 40.06
library(parallelsugar)
##
## Attaching package: ‘parallelsugar’
##
## The following object is masked from ‘package:parallel’:
##
## mclapply
system.time( mclapply(1:4, function(xx){ Sys.sleep(10) }) )
## user system elapsed
## 0.04 0.08 12.98
Progress bar
pblapply
- pbapply: Adding Progress Bar to '*apply' Functions
- https://peter.solymos.org/pbapply/reference/pbapply.html
pbmcapply
pbmcapply: Tracking the Progress of Mc*pply with Progress Bar.
Note: mclapply can be interrupted by Ctrl+c, but pbmclapply does not take it.
Random seeds
future 1.19.1 - Making Sure Proper Random Numbers are Produced in Parallel Processing
How to set library path on a {parallel} R cluster
How to set library path on a {parallel} R cluster
snow package
http://cran.r-project.org/web/packages/snow/index.html
Supported cluster types are "SOCK", "PVM", "MPI", and "NWS".
multicore package
This package is removed from CRAN.
Consider using package ‘parallel’ instead.
foreach package
Changes in the foreach package version 1.4.8, Feb 2020.
Simple example
This package depends on one of the following
- doParallel - Foreach parallel adaptor for the parallel package
- doSNOW - Foreach parallel adaptor for the snow package
- doMC - Foreach parallel adaptor for the multicore package. Used in glmnet vignette.
- How to kill a doMC worker when it's done?. The doMC package is basically a wrapper around the mclapply function, and mclapply forks workers that should exit before it returns.
- doMPI - Foreach parallel adaptor for the Rmpi package
- doRedis - Foreach parallel adapter for the rredis package
as a backend.
library(foreach) library(doParallel) m <- matrix(rnorm(9), 3, 3) cl <- makeCluster(2, type = "SOCK") registerDoParallel(cl) # register the parallel backend with the foreach package foreach(i=1:nrow(m), .combine=rbind) %dopar% (m[i,] / mean(m[i,])) stopCluster(cl)
See also this post Updates to the foreach package and its friends on Oct 2015.
combine list of lists
- .combine argument https://stackoverflow.com/questions/27279164/output-list-of-two-rbinded-data-frames-with-foreach-in-r
- Merge lists by mapply() or base::Map()
comb <- function(...) {
mapply('cbind', ..., SIMPLIFY=FALSE)
}
library(foreach)
library(doParallel)
cl <- makeCluster(2)
registerDoParallel(cl) # register the parallel backend with the foreach package
m <- rbind(rep(1,3), rep(2,3))
# nrow(m) can represents number of permutations (2 in this toy example)
tmp <- foreach(i=1:nrow(m)) %dopar% {
a <- m[i, ]
b <- a * 10
list(a, b)
}; tmp
# [[1]]
# [[1]][[1]]
# [1] 1 1 1
#
# [[1]][[2]]
# [1] 10 10 10
#
#
# [[2]]
# [[2]][[1]]
# [1] 2 2 2
#
# [[2]][[2]]
# [1] 20 20 20
foreach(i=1:nrow(m), .combine = "comb") %dopar% {
a <- m[i,]
b <- a * 10
list(a, b)
}
# [[1]]
# [,1] [,2]
# [1,] 1 2
# [2,] 1 2
# [3,] 1 2
#
# [[2]]
# [,1] [,2]
# [1,] 10 20
# [2,] 10 20
# [3,] 10 20
stopCluster(cl)
Replacing double loops
- https://stackoverflow.com/questions/30927693/how-can-i-parallelize-a-double-for-loop-in-r
- http://www.exegetic.biz/blog/2013/08/the-wonders-of-foreach/
library(foreach)
library(doParallel)
nc <- 4
nr <- 2
cores=detectCores()
cl <- makeCluster(cores[1]-1)
registerDoParallel(cl)
# set.seed(1234) # not work
# set.seed(1234, "L'Ecuyer-CMRG") # not work either
# library("doRNG")
# registerDoRNG(seed = 1985) # not work with nested foreach
# Error in list(e1 = list(args = (1:nr)(), argnames = "i", evalenv = <environment>, :
# nested/conditional foreach loops are not supported yet.
m <- foreach (i = 1:nr, .combine='rbind') %:% # nesting operator
foreach (j = 1:nc) %dopar% {
rnorm(1, i*5, j) # code to parallelise
}
m
stopCluster(cl)
Note that since the random seed (see the next session) does not work on nested loop, it is better to convert nested loop (two indices) to a single loop (one index).
set seed and doRNG package
- Vignette, Documentation
- doRNG package example
- How to set seed for random simulations with foreach and doMC packages?
- Use clusterSetRNGStream() from the parallel package; see How-to go parallel in R – basics + tips
- http://www.stat.colostate.edu/~scharfh/CSP_parallel/handouts/foreach_handout.html#random-numbers
library("doRNG") # doRNG does not need to be loaded after doParallel library("doParallel") cl <- makeCluster(2) registerDoParallel(cl) registerDoRNG(seed = 1234) # works for a single loop m1 <- foreach(i = 1:5, .combine = 'c') %dopar% rnorm(1) registerDoRNG(seed = 1234) m2 <- foreach(i = 1:5, .combine = 'c') %dopar% rnorm(1) identical(m1, m2) stopCluster(cl) attr(m1, "rng") <- NULL # remove rng attribute - Another way to use the seed is to supply .options.RNG in foreach() function.
r1 <- foreach(i=1:4, .options.RNG=1234) %dorng% { runif(1) }
Export libraries, variables, functions
clusterEvalQ(cl, {
library(biospear)
library(glmnet)
library(survival)
})
clusterExport(cl, list("var1", "foo2"))
If we use makeCluster(, "FORK"), then we don't need to do these.
Summary the result
foreach returns the result in a list. For example, if each component is a matrix we can use
- Reduce("+", res)/length(res) # Reduce("+", res, na.rm = TRUE) not working
- apply(simplify2array(res), 1:2, mean, na.rm = TRUE)
to get the average of matrices over the list.
Read a list files
Reading List Faster With parallel, doParallel, and pbapply
Don't use too many cores: Error in unserialize(socklist[[n]])
error reading from connection. See https://stackoverflow.com/a/24352032
Why is foreach() sometimes slower than for?
See https://stackoverflow.com/a/10414280 or https://stackoverflow.com/q/16963808
- foreach is only advisable if you have relatively few rounds through very time consuming functions.
- foreach() is best when the number of jobs does not hugely exceed the number of processors you will be using.
library(foreach) library(parallel) nCores <- future::availableCores() cl <- makeCluster(nCores[[1]]) registerDoParallel(cl) B<-10000 myFunc<-function() for(i in 1:B) sqrt(i) myFunc2<-function() foreach(i = 1:B) %do% sqrt(i) myParFunc<-function() foreach(i = 1:B) %dopar% sqrt(i) system.time(myFunc()) # user system elapsed # 0.001 0.002 0.004 system.time(myFunc2()) # user system elapsed # 2.473 0.005 2.477 system.time(myParFunc()) # user system elapsed # 3.560 0.242 3.829 stopCluster(cl)
Another example: reduce the probe sets corresponding to the same gene symbol by taking the probe set with the largest gene expression. It is not just the speed problem; it runs out of memory on a 44928x393 matrix with a PC has 8GB RAM. On a smaller data with 5000 genes and 100 arrays it took 0.071 seconds using split() + sapply() but it took 39 seconds using the foreach method.
require(magrittr) x <- matrix(rnorm(5000*100), nr=5000) # gene expression y <- sample(3000, 5000, replace = TRUE) # gene symbol rownames(x) <- names(y) <- 1:5000 # probe sets name spt <- apply(x,1,mean,na.rm=TRUE) %>% split(., y) ind <- sapply(spt, function(a) names(which.max(a))) x.reduced <- x[ind, ]; rownames(x.reduced) <- names(spt)
Progress bar (doSNOW + foreach)
- How to show the progress of code in parallel computation in R? It looks great.
| doParallel | doSNOW |
|---|---|
require(doParallel)
cores <- parallel::detectCores()
cl <- makeCluster(cores)
registerDoParallel(cl)
opts <- list(progress=progress)
result <- foreach(i=1:N) %dopar% {}
stopCluster(cl)
|
require(doSNOW)
require(Kendall)
cores <- parallel::detectCores()
cl <- makeSOCKcluster(cores)
registerDoSNOW(cl)
mydata <- matrix(rnorm(8000*500), ncol=500)
pb <- txtProgressBar(min=1, max=8000, style=3)
progress <- function(n) setTxtProgressBar(pb, n)
opts <- list(progress=progress)
result <- foreach(i=1:8000, .packages="Kendall",
.options.snow=opts, .combine='rbind') %dopar% {
abc <- MannKendall(mydata[i,])
data.frame(tau=abc$tau, sl=abc$sl)
}
close(pb)
stopCluster(cl)
|
- Monitoring progress of a foreach parallel job It also works on doSNOW but not doParallel backend.
snowfall package
Rmpi package
Some examples/tutorials
- http://trac.nchc.org.tw/grid/wiki/R-MPI_Install
- http://www.arc.vt.edu/resources/software/r/index.php
- https://www.sharcnet.ca/help/index.php/Using_R_and_MPI
- http://math.acadiau.ca/ACMMaC/Rmpi/examples.html
- http://www.umbc.edu/hpcf/resources-tara/how-to-run-R.html
- Ryan Rosario
- http://pj.freefaculty.org/guides/Rcourse/parallel-1/parallel-1.pdf
- * http://biowulf.nih.gov/apps/R.html
OpenMP
- R and openMP: boosting compiled code on multi-core cpu-s from parallelr.com.
BiocParallel
- BiocParallel bplapply()
- https://hpc.nih.gov/apps/R.html#biocparallel
- Orchestrating a small, parallel, RNA-seq pre-processing workflow using R
- DESeq2 Speed-up and parallelization thoughts
- Why DESeq2 in parallel mode is slower than normal?! Why the time does not increase when my computer has only 4 cores?
library("BiocParallel") register(SerialParam()) system.time({ bplapply(1:4, function(i) Sys.sleep(5)) }) # user system elapsed # 0.011 0.000 20.014 register(MulticoreParam(workers=4)) system.time({ bplapply(1:4, function(i) Sys.sleep(5)) }) # user system elapsed # 0.080 0.070 5.065 register(MulticoreParam(workers=8)) system.time({ bplapply(1:8, function(i) Sys.sleep(5)) }) # user system elapsed # 0.120 0.143 5.060 register(MulticoreParam(workers=16)) system.time({ bplapply(1:16, function(i) Sys.sleep(5)) }) # user system elapsed # 0.262 0.267 5.122 register(MulticoreParam(workers=32)) system.time({ bplapply(1:32, function(i) Sys.sleep(5)) }) # user system elapsed # 0.464 0.613 5.213 - It takes a so long time to run DESeq2 on 1000 samples. With 1000 sampes I would simply use limma-voom, see for references.
- DESeq2 using many cores despite parallel = FALSE you could try trace(bplapply)
RcppParallel
future & future.apply & doFuture packages
- https://cran.r-project.org/web/packages/future/index.html
- Asynchronous programming
- Parallelize Any Base R Apply Function
- Parallelize a For-Loop by Rewriting it as an Lapply Call
- Use foreach with HPC schedulers thanks to the future package
- useR! 2019 Slides on Futures
- future 1.15.0 - Lazy Futures are Now Launched if Queried
- rstudio::conf 2020 Slides on Futures
- Article 'A Unifying Framework for Parallel and Distributed Processing in R using Futures'
- Trust the Future
- future 1.20.1 - The Future Just Got a Bit Brighter
- Slides & video NYC R Meetup 2020.
- European Bioconductor Meeting 2020
- Packages importing future: Seurat
- functions have been written to take advantage of the future framework
- plan(multiprocess, workers = n) is deprecated. Read the NEWS. So be careful if we are following Parallelization in Seurat with future.
- normalizeData.XXX code example. future_lapply() was used (data was splitted into chunks). split() + future_lapply() + unlist()/do.call()
- Using Kubernetes and the Future Package to Easily Parallelize R in the Cloud
- Setting Up A Cluster of Tiny PCs For Parallel Computing – A Note To Myself 2026
furrr
furrr: Apply Mapping Functions in Parallel using Futures
txtProgressBar(), setTxtProgressBar()
Running R code for all combinations of some parameters
Running R code for all combinations of some parameters with lapply karate
Distributed programming: ddR, multidplyr
The evolution of distributed programming in R
Apache Spark
- Introduction to Apache Spark
- Mastering Apache Spark with R Javier Luraschi, Kevin Kuo, Edgar Ruiz (ebook)
Microsoft R Server
- Microsoft R Server (not Microsoft R Open)
GPU
- GPU Programming for All with ‘gpuR from parallelr.com. The gpuR is available on CRAN.
- gputools
Threads
- Rdsm package
- (A Very) Experimental Threading in R and a post from Mad Scientist
Benchmark
R functions to run timing
# Method 1 system.time( invisible(rnorm(10000))) # Method 2 btime <- Sys.time() invisible(rnorm(10000)) Sys.time() - btime # difftime(Sys.time(), btime)