Biowulf: Difference between revisions
| (60 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
[[File:Swarm fig 1.png|200px]] | [[File:Swarm fig 1.png|200px]] | ||
= Status = | |||
https://hpc.nih.gov/systems/status/ | |||
= helix and data transfer = | = helix and data transfer = | ||
* Data transfer and intensive file management tasks should not be performed on the Biowulf login node, biowulf.nih.gov. Instead such tasks should be performed on the dedicated interactive data transfer node (DTN), helix.nih.gov | * Data transfer and intensive file management tasks should not be performed on the Biowulf login node, biowulf.nih.gov. Instead such tasks should be performed on the dedicated interactive data transfer node (DTN), helix.nih.gov | ||
| Line 23: | Line 26: | ||
CentOS release 6.6 (Final) | CentOS release 6.6 (Final) | ||
</syntaxhighlight> | </syntaxhighlight> | ||
RHEL8/Rocky8 in June 2023. | |||
= Training notes = | = Training notes = | ||
| Line 35: | Line 39: | ||
== /scratch and [https://hpc.nih.gov/docs/userguide.html#local /lscratch] == | == /scratch and [https://hpc.nih.gov/docs/userguide.html#local /lscratch] == | ||
(Aug 2021). The /scratch directory will no longer be available from '''compute nodes''' (vs login node) effective Sep 1. | |||
* /scratch will continue to be accessible from Biowulf, Helix, HPCdrive and Globus. As before, /scratch can be used for temporary files that are expected to be automatically deleted in 10 days time. | |||
* Any batch or swarm command files which reference the /scratch file system should be modified before Sep 1, to use either your /data directory or /lscratch as appropriate. | |||
* https://hpc.nih.gov/storage/index.html | |||
The /scratch area on Biowulf is a large, low-performance '''shared area''' meant for the storage of temporary files. | The /scratch area on Biowulf is a large, low-performance '''shared area''' meant for the storage of temporary files. | ||
| Line 57: | Line 65: | ||
</pre> | </pre> | ||
== Transfer files == | == Transfer files (including large files) == | ||
* https://hpc.nih.gov/docs/transfer.html | * https://hpc.nih.gov/docs/transfer.html | ||
* [https://hpc.nih.gov/training/handouts/Data_Management_for_Groups.pdf#page=69 Data Management: Best Practices for Groups] | * [https://hpc.nih.gov/training/handouts/Data_Management_for_Groups.pdf#page=69 Data Management: Best Practices for Groups] | ||
* [https://hpc.nih.gov/docs/helixdrive.html Locally Mounting HPC System Directories] using [https://en.wikipedia.org/wiki/Server_Message_Block CIFS or SMB] protocol. This is simple. We just need to enter the password each time we connect to the server. | * [https://hpc.nih.gov/docs/helixdrive.html Locally Mounting HPC System Directories] using [https://en.wikipedia.org/wiki/Server_Message_Block CIFS or SMB] protocol. This is simple. We just need to enter the password each time we connect to the server. | ||
* [https://hpc.nih.gov/storage/globus.html Globus] | * [https://hpc.nih.gov/storage/globus.html Globus]. It is a web app to transfer files between local and remote machines. | ||
** https://www.globus.org/, | |||
** [https://www.globusworld.org/tour/program?c=32 GlobusWorld Tour Agenda] | |||
** [https://videocast.nih.gov/watch=52238 Day 1 recording] | |||
** [https://videocast.nih.gov/watch=52240 Day 2 recording] | |||
Alternatives: | Alternatives: | ||
* To transfer or share large amounts of data to specific individuals, located either at the NIH or at another institution, the HPC staff recommends | * To transfer or share '''large''' amounts of data to specific individuals, located either at the NIH or at another institution, the HPC staff recommends [https://hpc.nih.gov/storage/globus.html Globus]. | ||
* To share files with other users within NIH, CIT provides OneDrive - see https://myitsm.nih.gov/sp?id=kb_article&sys_id=1e424bebdbbc2f80dcd074821f9619c4 for details. | * To share files with other users within NIH, CIT provides OneDrive - see https://myitsm.nih.gov/sp?id=kb_article&sys_id=1e424bebdbbc2f80dcd074821f9619c4 for details. | ||
* To share small amounts of data with external or NIH colleagues, NIH makes the Box service (https://nih.account.box.com/login) available. You will need to request a Box account, if you do not already have one. It is possible to transfer data directly to and from the HPC systems using Box - see https://hpc.nih.gov/docs/box.html for details. | * To share small amounts of data with external or NIH colleagues, NIH makes the Box service (https://nih.account.box.com/login) available. You will need to request a Box account, if you do not already have one. It is possible to transfer data directly to and from the HPC systems using Box - see https://hpc.nih.gov/docs/box.html for details. | ||
| Line 72: | Line 84: | ||
== Create a shared folder == | == Create a shared folder == | ||
See [[Linux#Uppercase_S_in_permissions_of_a_folder_and_setGID|Uppercase S in permissions of a folder and setGID]]. '''chmod -R 2770 ShareFolder'''. | * [https://hpc.nih.gov/docs/groups.html User Groups & Shared Directories on Biowulf/Helix] | ||
* Under certain circumstances, permissions and group ownership may be set incorrectly. In these cases, permissions can be set with the command '''chmod g+rw''' (for files) or '''chmod g+rwx''' (for directories), and group ownership can be set using '''chgrp GROUPNAME'''. | |||
* By default, subdirectories created within '''/data/GROUPNAME''' will NOT be writable by anyone other than the user who created it, although each group member will be able to read files within. To automatically allow members of the group write access to subdirectories created, EACH GROUP MEMBER must add the command '''umask 007''' to their '''~/.bashrc''' file. (Remember, with umask 007, newly created files and directories will have all permissions (read, write, and execute) for the user and the group, but no permissions for others. This is a good practice when you want to share data with other users in the same group but want to completely exclude users who are not group members). | |||
* See [[Linux#Uppercase_S_in_permissions_of_a_folder_and_setGID|Uppercase S in permissions of a folder and setGID]]. '''chmod -R 2770 ShareFolder'''. | |||
== Numeric number for a folder's owner == | |||
It could be the user (UID) account has been deleted. | |||
== Local disk and temporary files == | == Local disk and temporary files == | ||
| Line 181: | Line 199: | ||
== [https://hpc.nih.gov/docs/userguide.html#partitions Partition and freen] == | == [https://hpc.nih.gov/docs/userguide.html#partitions Partition and freen] == | ||
* https://hpc.nih.gov/docs/b2-userguide.html#cluster_status | * [https://hpc.nih.gov/docs/b2-userguide.html#cluster_status Cluster status] | ||
* [https://hpc.nih.gov/docs/biowulf_tools.html Biowulf Utilities] about '''freen''' | |||
* In the script file, we can use '''$SLURM_CPUS_PER_TASK''' to represent the number of cpus | * In the script file, we can use '''$SLURM_CPUS_PER_TASK''' to represent the number of cpus | ||
* In the swarm command, we can use '''-t''' to specify the threads and '''-g''' to specify the memory (GB). | * In the swarm command, we can use '''-t''' to specify the threads and '''-g''' to specify the memory (GB). | ||
| Line 395: | Line 414: | ||
forgo 5760 1-00:00:00 3-00:00:00 | forgo 5760 1-00:00:00 3-00:00:00 | ||
</pre> | </pre> | ||
== Quick jobs == | |||
Use the '''--partition=norm,quick''' option when you submit a job that requires < 4 hours; see [https://hpc.nih.gov/docs/userguide.html#partitions User Guide]. | |||
= [https://hpc.nih.gov/docs/userguide.html#int Interactive debugging] = | = [https://hpc.nih.gov/docs/userguide.html#int Interactive debugging] = | ||
| Line 413: | Line 435: | ||
If we have an interactive job on a certain node, we can directly ssh into that node to check for example the ''/lscratch/$SLURM_JOBID'' usage. The JobID can be obtained from the '''jobload''' command. | If we have an interactive job on a certain node, we can directly ssh into that node to check for example the ''/lscratch/$SLURM_JOBID'' usage. The JobID can be obtained from the '''jobload''' command. | ||
It also helps to modify ".ssh/config" to include Host cn* lines. See [https://hpc.nih.gov/apps/vscode.html VS Code on Biowulf]. | It also helps to modify ".ssh/config" to include Host cn* lines. See [https://hpc.nih.gov/apps/vscode.html VS Code on Biowulf] and [https://www.backarapper.com/add-ssh-keys-to-ssh-agent-on-startup-in-macos/ How to Add SSH-Keys to SSH-Agent on Startup in MacOS Operating System]. | ||
We can also use '''freen -n''' to see some properties of the node. | We can also use '''freen -n''' to see some properties of the node. | ||
| Line 419: | Line 441: | ||
= [https://hpc.nih.gov/docs/userguide.html#par Parallel jobs] = | = [https://hpc.nih.gov/docs/userguide.html#par Parallel jobs] = | ||
Parallel (MPI) jobs that run on more than 1 node: Use the environment variable $SLURM_NTASKS within the script to specify the number of MPI processes. | Parallel (MPI) jobs that run on more than 1 node: Use the environment variable $SLURM_NTASKS within the script to specify the number of MPI processes. | ||
== Memory and CPUs == | |||
Many nodes has 28 cpus and 256GB memory. The ''freen'' command shows 247g for these 256GB node. Luckily in my application, the run time does not change much after I allocate >= 20 CPUs and I don't need that large amount of memory. | |||
= Reproduciblity/Pipeline = | = Reproduciblity/Pipeline = | ||
== | == Singularity == | ||
<ul> | |||
* [https://ncihub.org/groups/itcr/cwig NCI Containers and Workflows Interest Group] | <li>(obsolete) http://singularity.lbl.gov/, (new) https://sylabs.io/singularity/, (github release) https://github.com/sylabs/singularity, <s>https://apptainer.org/documentation/</s> | ||
<li> https://hpc.nih.gov/apps/singularity.html | |||
* .sif (Singularity Image File) is an image file, not a definition file (*.def) | |||
* sif file is created using the ''sudo singularity build'' command on a system where you have sudo or root access. Below is an example of my_image.def. | |||
:<syntaxhighlight lang='sh'> | |||
Bootstrap: docker | |||
From: ubuntu:20.04 | |||
%post | |||
apt-get update && apt-get install -y python3 | |||
</syntaxhighlight> | |||
:Build the Singularity image (it looks better to use an extension name for easy management later): | |||
:<syntaxhighlight lang='sh'> | |||
sudo singularity build my_image.sif my_image.def | |||
sudo singularity build my_image.simg my_image.def | |||
sudo singularity build my_image my_image.def | |||
</syntaxhighlight> | |||
* The definition file is a text file. It does not have to have an extension name. This is like a Dockerfile used by Docker compose. | |||
* Cache images are located in $HOME/.singularity/cache by default. But we can modify it by | |||
:<syntaxhighlight lang='sh'> | |||
export SINGULARITY_CACHEDIR=/data/${USER}/.singularity | |||
</syntaxhighlight> | |||
* '''singularity cache list -v''' cannot return the image names. [https://stackoverflow.com/questions/68510166/singularity-equivalent-to-docker-image-list Singularity equivalent to "docker image list"]. | |||
* "singularity shell --bind /data/$USER,/fdb,/lscratch my-container.sif" will start a container | |||
* In Singularity, if you don’t specify the destination path in the container when using the --bind option, the destination path will be set equal to the source path. See [https://docs.sylabs.io/guides/3.0/user-guide/bind_paths_and_mounts.html Bind Paths and Mounts] from Singularity User Guide. | |||
* Normally we use the '''singularity shell''' or '''singularity run''' command to use a singularity image. However, a singularity file can be made so the created image file works like an '''executable/binary''' file. See the "rnaseq.def" example in Biowulf. PS: the definition file on Biowulf gave errors. A modified working one is available on [https://gist.github.com/arraytools/fa6352953678e4182920fd4eb50e1cfd Github]. Use the bioconda instead of the conda channel. See [https://www.protocols.io/view/installation-instructions-for-rna-seq-analysis-usi-bgffjtjn.pdf Installation instructions for RNA-seq analysis using a conda environment V.1]. | |||
:<syntaxhighlight lang='sh'> | |||
$ ./rnaseq3 cat /etc/os-release | |||
PRETTY_NAME="Debian GNU/Linux 11 (bullseye)" <- Container | |||
$ cat /etc/os-release | |||
PRETTY_NAME="Ubuntu 22.04.4 LTS" <- Host | |||
</syntaxhighlight> | |||
<li>Install from source (in order to obtain a certain version of Singularity). [https://docs.sylabs.io/guides/4.1/admin-guide/admin_quickstart.html#installation-from-source V4.1]. It works on Debian 12. | |||
<li>[https://ncihub.org/groups/itcr/cwig NCI Containers and Workflows Interest Group] | |||
<li>[[R#R_and_Singularity|R and Singularity]] | |||
<li>[https://hpc.nih.gov/training/index.php Building a reproducible workflow with Snakemake and Singularity] | |||
</ul> | |||
=== Definition file === | |||
* [https://docs.sylabs.io/guides/4.1/user-guide/definition_files.html Definition files] from the current version V4.1. | |||
** [https://docs.sylabs.io/guides/4.1/user-guide/definition_files.html#header Header] and bootstrap agents | |||
** [https://docs.sylabs.io/guides/4.1/user-guide/definition_files.html#sections Sections]: '''%setup, %files, %environment, %post, %runscript, %startscript, %test, %labels, %help''' | |||
** [https://docs.sylabs.io/guides/4.1/user-guide/definition_files.html#scif-apps SCIF Apps] allows to encapsulate multiple apps into a container. '''%apprun, %applabels, %appinstall, %appenv, %apphelp''' | |||
::<syntaxhighlight lang='sh'> | |||
$ singularity exec --app app1 my_container.sif | |||
$ singularity exec --app app2 my_container.sif | |||
</syntaxhighlight> | |||
* Does the definition file depend on Singularity version? | |||
** Yes, the Singularity definition file can depend on the version of Singularity you are using. Different versions of Singularity may support different features, options, and syntax in the definition file. | |||
** For example, some options or features that are available in newer versions of Singularity might not be available in older versions. Conversely, some options or syntax that were used in older versions of Singularity might be deprecated or changed in newer versions. | |||
* [https://www.youtube.com/watch?v=m8llDjFuXlc Singularity Example Workflow] | |||
=== Import docker images === | |||
[https://docs.sylabs.io/guides/4.1/user-guide/singularity_and_docker.html#building-from-docker-oci-containers Building From Docker / OCI Containers -> Archives & Docker Daemon -> Containers in Docker Archive Files] | |||
<syntaxhighlight lang='sh'> | |||
# Step 1: create a docker archive file | |||
docker save -o dockerimage.tar <image_name> | |||
# Step 2: convert docker image to singularity image | |||
singularity build myimage.sif docker-archive://dockerimage.tar | |||
</syntaxhighlight> | |||
=== exec, run, shell === | |||
* '''singularity exec <container> <command>''': This command allows you to execute a specific command (required) inside a Singularity container. It starts a container based on the specified image and runs the '''command''' provided on the command line following singularity exec <image file name>. This will override any default command specified within the image metadata that would otherwise be run if you used singularity run. | |||
* '''singularity run''': This command runs the '''%runscript''' section of the Singularity definition file. If you have specified a command in the %runscript section (like starting the httpgd server), this command will be executed. This is useful when you want to use the Singularity image to perform a specific task or run a specific service. | |||
* '''singularity shell''': This command starts an interactive '''shell''' session inside the Singularity container. You can use this command to explore the file system of the Singularity image, install additional packages, or run commands interactively. This is useful for debugging or when you want to interactively work with the software installed in the Singularity image. | |||
Tips: | |||
* it seems "singularity run" and "singularity shell" do not expect any command as an argument. | |||
* if we specify %runscript in a definition, we can use the image by either | |||
** singularity exec XXX.sif /bin/bash # will ignore the %runscript and call the command | |||
** singularity run XXX.sif # will use the %runscript section | |||
** singularity shell XXX.sif # will ignore the %runscript section and launch a shell (/bin/sh) | |||
=== Bioconductor + httpgd === | |||
See [[Docker#R_and_httpgd_package|Docker -> R and httpgd package]]. | |||
For some reason, I see the port is open from the netstat command but FF cannot access it. "curl localhost:8888" showed the detail. | |||
=== Running services === | |||
https://docs.sylabs.io/guides/4.1/user-guide/running_services.html | |||
:<syntaxhighlight lang='sh'> | |||
singularity pull library://alpine | |||
# Starting Instances | |||
singularity instance start alpine_latest.sif instance1 | |||
singularity instance list | |||
singularity instance start alpine_latest.sif instance2 | |||
# Interact with instances | |||
singularity exec instance://instance1 cat /etc/os-release | |||
singularity run instance://instance2 # run the %runscript for the container | |||
singularity shell instance://instance2 | |||
== | # Stopping Instances | ||
* https://snakemake.readthedocs.io/en/stable/ | singularity instance stop instance1 | ||
singularity instance stop --all | |||
# PS. if an instance was launched by "sudo", we need to use | |||
# "sudo" to `list` and `stop` the instance. | |||
# “Hello-world” | |||
sudo singularity build nginx.sif nginx.def | |||
sudo singularity instance start --writable-tmpfs nginx.sif web | |||
# INFO: instance started successfully | |||
curl localhost | |||
sudo singularity instance stop web | |||
</syntaxhighlight> | |||
Check out the documentation for "API Server Example" where port 9000 was used. In that case, we do not need to use "sudo" to run the service. Below is a working example of running nginx on a non-default port. First, create a file '''default.conf'''. | |||
<pre> | |||
events { | |||
worker_connections 1024; | |||
} | |||
http { | |||
server { | |||
listen 8080; | |||
location / { | |||
root /usr/share/nginx/html; | |||
index index.html index.htm; | |||
} | |||
} | |||
} | |||
</pre> | |||
Second, create a definition file '''nginx.def'''. | |||
{{Pre}} | |||
Bootstrap: docker | |||
From: nginx | |||
%startscript | |||
nginx -c /etc/nginx/conf.d/default.conf | |||
%files | |||
/home/brb/singularity/nginx/default.conf /etc/nginx/conf.d/default.conf | |||
</pre> | |||
Last, build the image and run the service. | |||
<pre> | |||
sudo singularity build nginx.sif nginx.def | |||
sudo singularity instance start --writable-tmpfs nginx.sif web2 | |||
curl localhost:8080 | |||
sudo singularity instance stop -a | |||
</pre> | |||
=== Applications === | |||
* [https://zavolanlab.github.io/zarp-cli/guides/installation/ ZARP-cli] & [https://github.com/zavolanlab/zarp ZARP] for Automated RNA-seq Pipeline. See [https://www.rna-seqblog.com/rna-seq-analysis-made-easy/ RNA-Seq analysis made easy] | |||
=== Singularity vs Podman === | |||
<ul> | |||
<li>Singularity vs Podman - Podman and Singularity have different strengths and are suited for different use cases. Podman is more similar to Docker and is suitable for general-purpose containerization, while Singularity is more specialized for use in HPC and scientific computing environments. | |||
* Rootless Containers: | |||
** Podman: Podman allows for running containers as non-root users, which can be useful for security and isolation. | |||
** Singularity: Singularity also supports running containers as non-root users, but it is primarily designed for environments where users do not have root access, such as HPC systems. | |||
* Image Building: | |||
** Podman: Podman has built-in support for building images using a Dockerfile or OCI (Open Container Initiative) image specification. | |||
** Singularity: Singularity uses a different image format and requires a Singularity definition file to build images, which can be '''more complex than using a Dockerfile'''. | |||
* Image Distribution: | |||
** Podman: Podman can pull and push images from and to various container registries, similar to Docker. | |||
** Singularity: Singularity images are typically distributed as single files (.sif files) and can be '''more easily shared and moved between systems'''. | |||
* User Experience: | |||
**Podman: Podman is designed to have a command-line interface similar to Docker, making it easy for Docker users to transition to Podman. | |||
** Singularity: Singularity has its own set of commands and is designed with a focus on compatibility with existing HPC workflows and environments. | |||
* Use Cases: | |||
** Podman: Podman is suitable for a wide range of use cases, including development, testing, and production environments. | |||
** Singularity: Singularity is more commonly used in HPC and scientific computing environments, where users need to run containers on shared systems without root access. | |||
<li>Why Singularity is better than Podman? Singularity has some features that are not available in Podman, which make it particularly well-suited for certain use cases, especially in high-performance computing (HPC) environments: | |||
* '''Compatibility with HPC Environments''': Singularity is designed to work seamlessly in HPC environments where users often lack administrative privileges and need to run containers on shared systems. Its ability to run containers without requiring root access and its support for common HPC features make it a popular choice in these environments. | |||
* '''Secure Execution''': Singularity's security model, which runs containers with the same permissions as the user who launched them, provides a level of security and isolation that is well-suited for environments where security is a concern. | |||
* '''Image Format''': Singularity uses a different image format (.sif files) than Podman (OCI image format). Singularity images are typically distributed as single files, which can be ''more convenient for sharing and moving between systems, especially in environments where network access is limited''. | |||
** <span style="color: brown">A singularity image is portable across platforms, like Snap/Flatpak/AppImage in Linux. For example, if I use "singularity build r-base docker://r-base" to download the latest R image on a Rocky linux (host OS), then the image can be directly run on a Ubuntu linux (host OS).</span> | |||
* '''Environment Preservation''': ''Singularity is designed to preserve the user's environment inside the container, including environment variables, file system mounts, and user identity. This makes it easier to replicate the user's environment inside the container, which can be important in HPC and scientific computing workflows.'' | |||
** <span style="color: brown">For the example of an r-base image, an X11 window will open when I draw a plot inside a singularity container.</span> | |||
<li>Why Podman is better than Singularity? Podman has some features that are not available in Singularity, which make it particularly useful for certain use cases: | |||
* '''Rootless Containers''': Podman allows users to run containers as non-root users, providing a level of security and isolation that is useful in multi-user environments. Singularity also supports running containers as non-root users, but Podman's approach may be more flexible and easier to use in some cases. | |||
* '''Buildah Integration''': Podman integrates with Buildah, a tool for building OCI (Open Container Initiative) container images. This integration allows users to build and manage container images using the same toolset, providing a seamless workflow for container development. | |||
* '''Docker Compatibility''': Podman is designed to be compatible with the Docker CLI (Command Line Interface), making it easy for Docker users to transition to Podman. This compatibility extends to Docker Compose, allowing users to use Podman to manage multi-container applications defined in Docker Compose files. | |||
* '''Container Management''': Podman provides a range of commands for managing containers, including starting, stopping, and removing containers, as well as inspecting container logs and stats. While Singularity also provides similar functionality, Podman's CLI may be more familiar to users accustomed to Docker. | |||
* '''Networking''': Podman provides advanced networking features, including support for CNI (Container Network Interface) plugins, which allow users to configure custom networking solutions for their containers. Singularity's networking capabilities are more limited in comparison. | |||
</ul> | |||
== Snakemake and python == | |||
<ul> | |||
<li>Snakemake is a workflow management system that aims to reduce the complexity of creating workflows by providing a fast and comfortable execution environment, together with a clean and modern specification language in python style. | |||
<li>https://snakemake.readthedocs.io/en/stable/ | |||
<li>https://hpc.nih.gov/apps/snakemake.html | |||
<li>[https://bioinformaticsreview.com/20211123/installing-snakemake-on-ubuntu-linux/ Install on Ubuntu]. Before installing Snakemake, you need to | |||
* [https://bioinformaticsreview.com/20211116/installing-conda-on-ubuntu-linux/ Install Conda]. | |||
* Install Mamba | |||
:<syntaxhighlight lang='sh'> | |||
$ conda install mamba -n base -c conda-forge | |||
</syntaxhighlight> | |||
* Install snakemake | |||
:<syntaxhighlight lang='sh'> | |||
$ conda activate base | |||
$ mamba create -c conda-forge -c bioconda -n snakemake snakemake | |||
</syntaxhighlight> | |||
* Finally | |||
:<syntaxhighlight lang='sh'> | |||
$ conda activate snakemake | |||
$ snakemake --help | |||
</syntaxhighlight> | |||
</ul> | |||
= Apps = | = Apps = | ||
[https://hpc.nih.gov/apps/ Scientific Applications on NIH HPC Systems] | [https://hpc.nih.gov/apps/ Scientific Applications on NIH HPC Systems] | ||
= scp = | |||
Copy a file from helix to local. | |||
How to handle the path contains spaces. | |||
<pre> | |||
scp helix:"/data/xxx/ABC abc/file.txt" /tmp/ | |||
</pre> | |||
The double quotes also work for local directories/files. | |||
= R program = | = R program = | ||
| Line 442: | Line 673: | ||
</syntaxhighlight> | </syntaxhighlight> | ||
where '''-r''' means to use regular expression match. This will match "R/3.5.2" or "R/3.5" but not "Rstudio/1.1.447". | where '''-r''' means to use regular expression match. This will match "R/3.5.2" or "R/3.5" but not "Rstudio/1.1.447". | ||
== pacman == | |||
Biowulf website used the [https://cran.r-project.org/web/packages/pacman/index.html pacman] package to manage R packages. Interestingly, pacman is also a command used by [[Arch_Linux#Pacman pacman|Arch Linux]]. | |||
== (Self-installed) R package directory == | == (Self-installed) R package directory == | ||
| Line 448: | Line 682: | ||
The directory '''~/R/x86_64-pc-linux-gnu-library/''' was not used anymore in Biowulf. | The directory '''~/R/x86_64-pc-linux-gnu-library/''' was not used anymore in Biowulf. | ||
== RStudio == | <pre> | ||
> .libPaths() | |||
[1] "/usr/local/apps/R/4.2/site-library_4.2.0" | |||
[2] "/usr/local/apps/R/4.2/4.2.0/lib64/R/library" | |||
> packageVersion("Greg") | |||
Error in packageVersion("Greg") : there is no package called ‘Greg’ | |||
> install.packages("Greg") | |||
Installing package into ‘/usr/local/apps/R/4.2/site-library_4.2.0’ | |||
(as ‘lib’ is unspecified) | |||
Warning in install.packages("Greg") : | |||
'lib = "/usr/local/apps/R/4.2/site-library_4.2.0"' is not writable | |||
Would you like to use a personal library instead? (yes/No/cancel) yes | |||
Would you like to create a personal library | |||
‘/home/XXXXXX/R/4.2/library’ | |||
to install packages into? (yes/No/cancel) yes | |||
--- Please select a CRAN mirror for use in this session --- | |||
Secure CRAN mirrors | |||
1: 0-Cloud [https] | |||
... | |||
74: USA (IA) [https] | |||
75: USA (MI) [https] | |||
76: USA (OH) [https] | |||
77: USA (OR) [https] | |||
78: USA (TN) [https] | |||
79: USA (TX 1) [https] | |||
80: Uruguay [https] | |||
81: (other mirrors) | |||
Selection: 74 | |||
trying URL 'https://mirror.las.iastate.edu/CRAN/src/contrib/Greg_1.4.1.tar.gz' | |||
Content type 'application/x-gzip' length 205432 bytes (200 KB) | |||
> .libPaths() | |||
[1] "/spin1/home/linux/XXXXXX/R/4.2/library" | |||
[2] "/usr/local/apps/R/4.2/site-library_4.2.0" | |||
[3] "/usr/local/apps/R/4.2/4.2.0/lib64/R/library" | |||
</pre> | |||
== RStudio (standalone application) == | |||
This approach requires running the NoMachine app. | |||
<ul> | |||
<li>Step 1: [https://hpc.nih.gov/docs/nx.html Connecting to Biowulf with NoMachine (NX)] (Tip: no need to use ssh from a terminal) | |||
* It works though the resolution is not great on a Mac screen. This assume we are in the NIH network. | |||
* One thing is about adjusting the window size. Click '''Ctrl + Alt + 0''' to bring up the setting if we accept all the default options when we connect to a remote machine. Click 'Resize remote display'. Now we can use mouse to drag and adjust the display size. This will keep the resolution as it showed up originally. It is easier when I choose the 'key-based authorization method with a key you provide'. | |||
* Desktop environment is XFCE ($DESKTOP_SESSION) | |||
<li>Step 2: Use [https://hpc.nih.gov/apps/RStudio.html RStudio on Biowulf]. | |||
* We need to open a terminal app from the RedHat OS desktop and follow the instruction there. | |||
* The desktop IDE (NOT a web application) program is installed in /usr/local/apps/rstudio/rstudio-1.4.1103/. | |||
* Problems: The copy/paste keyboard shortcuts do not work even I checked the option "Grab the keyboard input". Current resolution: Use two ssh connections (one to run R, and another one to edit file). Open noMachine and use the desktop to view graph files (gv XXX.pdf OR change the default program to 'GNU GV Postscript/PDF Viewer' from the default 'LibreOffice Draw'). | |||
<pre> | <pre> | ||
sinteractive --mem=6g --gres=lscratch:5 | sinteractive --mem=6g --gres=lscratch:5 | ||
| Line 462: | Line 739: | ||
rstudio & | rstudio & | ||
</pre> | </pre> | ||
</ul> | |||
== httpgd: Web application == | |||
This approach just need a Terminal and a browser. [https://hpc.nih.gov/docs/tunneling/ SSH tunnel] is used. The '''port''' number is shown in the output of "sinteractive --tunnel" command. | |||
Refer to the instruction for the [https://hpc.nih.gov/apps/jupyter.html Jupyter] case. The key is the port number has to be passed to R when we call hgd() function. | |||
<ul> | |||
<li>Terminal 1 | |||
<syntaxhighlight lang='sh'> | |||
ssh biowulf.nih.gov | |||
sinteractive --gres=lscratch:5 --mem=10g --tunnel | |||
# Please create a SSH tunnel from your workstation to these ports on biowulf. | |||
# On Linux/MacOS, open a terminal and run: | |||
# | |||
# ssh -L 33327:localhost:33327 [email protected] | |||
module load R/4.3 | |||
R | |||
> library(httpgd) | |||
> httpgd::hgd(port = as.integer(Sys.getenv("PORT1"))) | |||
httpgd server running at: | |||
http://127.0.0.1:33327/live?token=FpaxiBKz | |||
</syntaxhighlight> | |||
<li>Terminal 2 | |||
<syntaxhighlight lang='sh'> | |||
ssh -L 33327:localhost:33327 biowulf.nih.gov | |||
</syntaxhighlight> | |||
Open the URL http://127.0.0.1:33327/live?token=FpaxiBKz given by httpgd::hgd() on my local browser. | |||
</ul> | |||
Call "exit" twice on Terminal 1 and "exit" once on Terminal 2 to quit Biowulf. | |||
= Python and conda = | |||
* https://hpc.nih.gov/apps/python.html. Search '''pip'''. | |||
* [https://hpc.nih.gov/apps/jupyter.html Jupyter on Biowulf] | |||
= Visual Studio Code = | = Visual Studio Code = | ||
| Line 480: | Line 791: | ||
== Jupyter == | == Jupyter == | ||
[https://hpc.nih.gov/apps/jupyter.html Jupyter on Biowulf] | [https://hpc.nih.gov/apps/jupyter.html Jupyter on Biowulf] | ||
The following diagram contains steps to launch Jupyter with an optional conda environment. | |||
[[File:Jupyter.drawio.png|300px]] | |||
= Terminal customization = | = Terminal customization = | ||
| Line 495: | Line 810: | ||
== tmux for keeping SSH sessions == | == tmux for keeping SSH sessions == | ||
[[Terminal_multiplexer#tmux.2A|tmux]] | [[Terminal_multiplexer#tmux.2A|tmux]] | ||
= Reference genomes = | |||
{{Pre}} | |||
$ wc -l /fdb/STAR/iGenomes/Homo_sapiens/UCSC/hg19/genes.gtf | |||
1006819 /fdb/STAR/iGenomes/Homo_sapiens/UCSC/hg19/genes.gtf | |||
$ wc -l /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf | |||
1006819 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf | |||
# Transcript | |||
$ grep uc002gig /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/* | |||
/fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/kgXref.txt:uc002gig.1 AM076971 E7EQX7 E7EQX7_HUMAN TP53 NM_001126114 NP_001119586 Homo sapiens tumor protein p53 (TP53), transcript variant 2, mRNA. | |||
/fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownGene.txt:uc002gig.1 chr17 - 7565096 7579937 75652567579912 7 7565096,7577498,7578176,7578370,7579311,7579699,7579838, 7565332,7577608,7578289,7578554,7579590,7579721,7579937, E7EQX7 uc002gig.1 | |||
/fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownToRefSeq.txt:uc002gig.1 NM_001126114 | |||
$ ls -logh /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/ | |||
total 231M | |||
-rwxrwxr-x 1 31K Jul 21 2015 cytoBand.txt | |||
-rwxrwxr-x 1 136M Jul 21 2015 genes.gtf | |||
-rwxrwxr-x 1 11M Jul 21 2015 kgXref.txt | |||
-rwxrwxr-x 1 20M Jul 21 2015 knownGene.txt | |||
-rwxrwxr-x 1 1.6M Jul 21 2015 knownToRefSeq.txt | |||
-rwxrwxr-x 1 4.6M Jul 21 2015 refFlat.txt.gz | |||
-rwxrwxr-x 1 15M Jul 26 2015 refGene.txt | |||
-rwxrwxr-x 1 45M Jul 21 2015 refSeqSummary.txt | |||
$ wc -l /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownGene.txt | |||
82960 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownGene.txt | |||
$ wc -l /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf | |||
1006819 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf | |||
$ head -4 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf | |||
chr1 unknown exon 11874 12227 . + . gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS16932"; | |||
chr1 unknown exon 12613 12721 . + . gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS16932"; | |||
chr1 unknown exon 13221 14409 . + . gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS16932"; | |||
chr1 unknown exon 14362 14829 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; | |||
$ head -4 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownGene.txt | |||
uc001aaa.3 chr1 + 11873 14409 11873 11873 3 11873,12612,13220, 12227,12721,14409, uc001aaa.3 | |||
uc010nxr.1 chr1 + 11873 14409 11873 11873 3 11873,12645,13220, 12227,12697,14409, uc010nxr.1 | |||
uc010nxq.1 chr1 + 11873 14409 12189 13639 3 11873,12594,13402, 12227,12721,14409, B7ZGX9 uc010nxq.1 | |||
uc009vis.3 chr1 - 14361 16765 14361 14361 4 14361,14969,15795,16606, 14829,15038,15942,16765, uc009vis.3 | |||
</pre> | |||
== Convert transcript to symbol == | |||
[[Anders2013#Convert_transcript_to_symbol|Anders2013 → here]] | |||
Latest revision as of 21:06, 31 August 2024
Status
https://hpc.nih.gov/systems/status/
helix and data transfer
- Data transfer and intensive file management tasks should not be performed on the Biowulf login node, biowulf.nih.gov. Instead such tasks should be performed on the dedicated interactive data transfer node (DTN), helix.nih.gov
- Examples of such tasks include:
- cp, mv, rm commands on large numbers of files or directories
- file compression/uncompression (tar, zip, etc.)
- file transfer (sftp, scp, rsync, etc.)
- https://helix.nih.gov/ Helix is an interactive system for short jobs. Moving large data transfers to Helix, which is now designated for interactive data transfers. For instance, use Helix when transferring hundreds of gigabytes of data or more using any of these commands: cp, scp, rsync, sftp, ascp, etc.
- https://hpc.nih.gov/docs/rhel7.html#helix Helix transitioned to becoming the dedicated interactive data transfer and file management node [1] and its hardware was later upgraded to support this role [2]. Running processes such as scp, rsync, tar, and gzip on the Biowulf login node has been discouraged ever since.
Linux distribution
$ ls /etc/*release # login mode $ cat /etc/redhat-release Red Hat Enterprise Linux Server release 6.8 (Santiago) $ sinteractive # switch to biowulf2 computing nodes $ cat /etc/redhat-release CentOS release 6.6 (Final) $ cat /etc/centos-release CentOS release 6.6 (Final)
RHEL8/Rocky8 in June 2023.
Training notes
- https://hpc.nih.gov/training/ Slides, Videos and Handouts from Previous HPC Classes
- https://hpc.nih.gov/docs/ Biowulf User Guides
- https://hpc.nih.gov/docs/B2training.pdf
- https://hpc.nih.gov/docs/biowulf2-beta-handout.pdf
- Online class: Introduction to Biowulf
Storage
/scratch and /lscratch
(Aug 2021). The /scratch directory will no longer be available from compute nodes (vs login node) effective Sep 1.
- /scratch will continue to be accessible from Biowulf, Helix, HPCdrive and Globus. As before, /scratch can be used for temporary files that are expected to be automatically deleted in 10 days time.
- Any batch or swarm command files which reference the /scratch file system should be modified before Sep 1, to use either your /data directory or /lscratch as appropriate.
- https://hpc.nih.gov/storage/index.html
The /scratch area on Biowulf is a large, low-performance shared area meant for the storage of temporary files.
- Each user can store up to a maximum of 10 TB in /scratch. However, 10 TB of space is not guaranteed to be available at any particular time.
- If the /scratch area is more than 80% full, the HPC staff will delete files as needed, even if they are less than 10 days old.
- Files in /scratch are automatically deleted 10 days after last access.
- Touching files to update their access times is inappropriate and the HPC staff will monitor for any such activity.
- Use /lscratch (not /scratch) when data is to be accessed from large numbers of compute nodes or large swarms.
- The central /scratch area should NEVER be used as a temporary directory for applications -- use /lscratch instead.
- fasterq-dump command in SRA-Toolkit where a temporary directory is needed. See HowTo page.
- Running RStudio interactively. It is generally recommended to allocate at least a small amount of lscratch for temporary storage for R.
- See the slides of Using the NIH HPC Storage Systems Effectively from NIH HPC classes
An example of using /lscratch space
fasterq-dump -t /lscratch/$SLURM_JOB_ID \ --split-files \ -O /lscratch/$SLURM_JOBID SRR13526458; \ pigz -p6 /lscratch/$SLURM_JOBID/SRR13526458*.fastq; \ cp /lscratch/$SLURM_JOBID/SRR13526458*.fastq.gz ~/PDACPDX
Transfer files (including large files)
- https://hpc.nih.gov/docs/transfer.html
- Data Management: Best Practices for Groups
- Locally Mounting HPC System Directories using CIFS or SMB protocol. This is simple. We just need to enter the password each time we connect to the server.
- Globus. It is a web app to transfer files between local and remote machines.
Alternatives:
- To transfer or share large amounts of data to specific individuals, located either at the NIH or at another institution, the HPC staff recommends Globus.
- To share files with other users within NIH, CIT provides OneDrive - see https://myitsm.nih.gov/sp?id=kb_article&sys_id=1e424bebdbbc2f80dcd074821f9619c4 for details.
- To share small amounts of data with external or NIH colleagues, NIH makes the Box service (https://nih.account.box.com/login) available. You will need to request a Box account, if you do not already have one. It is possible to transfer data directly to and from the HPC systems using Box - see https://hpc.nih.gov/docs/box.html for details.
User Dashboard
https://hpc.nih.gov/dashboard/
- User Groups & Shared Directories on Biowulf/Helix
- Under certain circumstances, permissions and group ownership may be set incorrectly. In these cases, permissions can be set with the command chmod g+rw (for files) or chmod g+rwx (for directories), and group ownership can be set using chgrp GROUPNAME.
- By default, subdirectories created within /data/GROUPNAME will NOT be writable by anyone other than the user who created it, although each group member will be able to read files within. To automatically allow members of the group write access to subdirectories created, EACH GROUP MEMBER must add the command umask 007 to their ~/.bashrc file. (Remember, with umask 007, newly created files and directories will have all permissions (read, write, and execute) for the user and the group, but no permissions for others. This is a good practice when you want to share data with other users in the same group but want to completely exclude users who are not group members).
- See Uppercase S in permissions of a folder and setGID. chmod -R 2770 ShareFolder.
Numeric number for a folder's owner
It could be the user (UID) account has been deleted.
Local disk and temporary files
See https://hpc.nih.gov/docs/b2-userguide.html#local and https://hpc.nih.gov/storage/
Dashboard
User dashboard Unlock account, disk usage, job info
Quota
checkquota
Environment modules
# What modules are available
module avail
module -d avail # default
module avail STAR
module spider bed # search by a case-insensitive keyword
# Load a module
module list # loaded modules
module load STAR
module load STAR/2.4.1a
module load plinkseq macs bowtie # load multiple modules
# if we try to load a module in a bash script, we can use the following
module load STAR || exit 1
# Unload a module
module unload STAR/2.4.1a
# Switch to a different version of an application
# If you load a module, then load another version of the same module, the first one will be unloaded.
# Examine a modulefile
$ module display STAR
-----------------------------------------------------------------------------
/usr/local/lmod/modulefiles/STAR/2.5.1b.lua:
-----------------------------------------------------------------------------
help([[This module sets up the environment for using STAR.
Index files can be found in /fdb/STAR
]])
whatis("STAR: ultrafast universal RNA-seq aligner")
whatis("Version: 2.5.1b")
prepend_path("PATH","/usr/local/apps/STAR/2.5.1b/bin")
# Set up personal modulefiles
# Using modules in scripts
# Shared Modules
Question: If I run "sinteractive" and "module load R" in one terminal (take note cnXXXX) and then run ssh cnXXXX in another terminal, the *R* instance is not available in the 2nd terminal. Why?
Single file - sbatch
- sbatch
- Note sbatch command does not support --module option. In sbatch case, the module command has to be put in the script file.
- The log file is slurm-XXXXXXXX.out
- If we want to create a script file, we can use the echo command; see Create a simple text file with multiple lines; write data to a file in bash script
- Script file must be starting with a line #!/bin/bash
Don't use the swarm command on a single script file since swarm will treat each line of the script file as an independent command.
sbatch --cpus-per-task=2 --mem=4g --time 24:00:00 MYSCRIPT # Use --time 24:00:00 to increase the wall time from the default 2 hours to 24 hours
An example of the script file (Slurm environment variable $SLURM_CPUS_PER_TASK within your script was used to specify the number of threads to the program)
#!/bin/bash module load novocraft novoalign -c $SLURM_CPUS_PER_TASK -f s_1_sequence.txt -d celegans -o SAM > out.sam
rslurm
rslurm package. Functions that simplify submitting R scripts to a Slurm workload manager, in part by automating the division of embarrassingly parallel calculations across cluster nodes.
Multiple files - swarm
- swarm. Remember the -f and --time options. The default walltime is 2 hours.
- Source code and other information on github
swarm -f run.sh --time 24:00:00 swarm -t 3 -g 20 -f run_seqtools_vc.sh --module samtools,picard,bwa --verbose 1 --devel # 3 commands run in 3 subjobs, each command requiring 20 gb and 3 threads, allocating 6 cores and 12 cpus swarm -t 3 -g 20 -f run_seqtools_vc.sh --module samtools,picard,bwa --verbose 1 # To change the default walltime, use --time 24:00:00 swarm -t 8 -g 24 --module tophat,samtools,htseq -f run_master.sh cat sw3n17156.o
Environment variables $SLURM
Swarm on Biowulf. Some examples: fmriprep, qsiprep.
- $SLURM_MEM_PER_NODE: 32768
- $SLURM_JOB_ID
- $SLURM_NNODES: 1
- $SLURM_CPUS_ON_NODE: 2
- $SLURM_JOB_CPUS_PER_NODE: 2
- $SLURM_NODELIST: cn2448
Why a job is pending
Partition and freen
- Cluster status
- Biowulf Utilities about freen
- In the script file, we can use $SLURM_CPUS_PER_TASK to represent the number of cpus
- In the swarm command, we can use -t to specify the threads and -g to specify the memory (GB).
Biowulf nodes are grouped into partitions. A partition can be specified when submitting a job. The default partition is 'norm'. The freen command can be used to see free nodes and CPUs, and available types of nodes on each partition.
We may need to run swarm commands on non-default partitions. For example, not many free CPUs are available in 'norm' partition. Or Total time for bundled commands is greater than partition walltime limit. Or because the default partition norm has nodes with a maximum of 120GB memory.
We can run the swarm command on different partition (the default is 'norm'). For example, to run on b1 parition (the hardware in b1 looks inferior to norm)
swarm -f outputs/run_seqtools_dge_align -g 20 -t 16 \
--module tophat,samtools,htseq \
--time 6:00:00 --partition b1 --verbose 1
If we want to restrict the output of freen to the norm nodes, we can use freen | grep -E 'Partition|----|norm' ; see the User Guide.
Partition FreeNds FreeCPUs FreeGPUs Cores CPUs GPUs Mem Disk Features ------------------------------------------------------------------------------------------------------- norm* 8 / 216 3702 / 14748 36 72 373g 3200g cpu72,core36,g384,ssd3200,x6140,ibhdr100 norm* 101 / 522 20030 / 29232 28 56 247g 800g cpu56,core28,g256,ssd800,x2680,ibfdr norm* 497 / 529 16696 / 16928 16 32 121g 800g cpu32,core16,g128,ssd800,x2650,10g norm* 504 / 539 29192 / 30184 28 56 247g 400g cpu56,core28,g256,ssd400,x2695,ibfdr norm* 6 / 7 342 / 392 28 56 247g 2400g cpu56,core28,g256,ssd2400,x2680,ibfdr
jobhist and track the resource usage
- jobhist XXXXXXXX will show the resource usage. The Jobid Runtime, MemUsed columns can be used to identify the job that was using most resource
- To identify the command run by a job, check out the log file <swarm_XXXXXX_Y.o>
Running R scripts
https://hpc.nih.gov/apps/R.html
Running a swarm of R batch jobs on Biowulf
$ cat Rjobs R --vanilla < /data/username/R/R1 > /data/username/R/R1.out R --vanilla < /data/username/R/R2 > /data/username/R/R2.out # swarm -g 32 -t 10 -f /home/username/Rjobs --module R/3.6.0 --time 6:00:00
Pay attention to the default wall time (eg 2 hours) and various swarm options. See Swarm.
Parallelizing with parallel.
- The following is modified from biowulf R's webpage. I change it so it works on non-biowulf system (like local Linux, Mac, or Windows).
- The number of allocated CPUs and available memory are related.
- Here I assume the Windows and Mac only has a modest RAM (eg 16GB) and the local Linux has enough RAM.
- The RAM size on the local system can be obtained through the commented lines.
detectBatchCPUs <- function() {
ncores <- as.integer(Sys.getenv("SLURM_CPUS_PER_TASK"))
if (is.na(ncores)) {
ncores <- as.integer(Sys.getenv("SLURM_JOB_CPUS_PER_NODE"))
}
if (is.na(ncores)) {
# the system is not biowulf
if (grepl("linux", R.version$os)) {
# if it is linux, we assume there are enough ram
# mem <- system('grep MemTotal /proc/meminfo', intern = TRUE)
# mem <- strsplit(mem, " ")[[1]]
# mem <- as.integer(mem[length(mem) -1])
ncores <- future::availableCores()
# } else if (grepl("darwin", R.version$os)) {
# ncores <- 2
} else ncores <- 2
}
return(ncores)
}
ncpus <- detectBatchCPUs()
options(mc.cores = ncpus)
mclapply(..., mc.cores = ncpus)
makeCluster(ncpus)
Some experiences
jobhist show significant less memory use than jobload. It appears that novoalign uses shared memory, which we account for in jobload but not in jobhist. In this case, you will want to use the memory value reported by jobload to avoid running out of allocated memory and thus getting killed by Slurm.
What is shared memory? Each process in Linux gets its own private application memory space. Linux makes available a portion of memory where multiple processes can also share the same memory space so these multiple processes can work on the same datasets.
Is there a way I can tell "shared memory" is in use when I was running a job? At the moment, you would have to be on a node running one of your jobs and run the 'ipcs -m' command. This lists shared memory segments for any processes using shared memory. This can be cumbersome, especially for a large multimode job or swarm.
freen command shows the maximum threads is 56 and the memory is 246GB.
When I run an R script (foreach is employed to loop over simulation runs), I find
- Assign 56 threads can guarantee 56 simulations run at the same time (check by the jobload command).
- We need to worry about the RAM size. The larger the threads, the more memory we need. If we don't assign enough memory, weird error message will be spit out.
- Even assigning 56 threads can help to run 56 simulations at the same time, the actual execution time is longer than when I run fewer simulations.
allocated threads allocated memory number of runs memory used time (min) 56 64 10 30 10 56 64 20 36 13 56 64 56 58 27
Monitor jobs/Delete/Kill jobs
Please don't run 'jobload' (or other slurm query commands) any more than once every 120 seconds. Running it every 30 sec puts an unnecessary load on the batch system.
sjobs watch -n 125 jobload watch -n 125 "jobload | tail" scancel -u XXXXX scancel NNNNN scancel --name=JobName scancel --state=PENDING scancel --state=RUNNING squeue -u XXXX jobhist 17500 # report the CPU and memory usage of completed jobs.
The other two commands are very useful too jobhist and swarmhist (temporary).
$ cat /usr/local/bin/swarmhist
#!/bin/bash
usage="usage: $0 jobid"
jobid=$1
[[ -n $jobid ]] || { echo $usage; exit 1; }
ret=$(grep "jobid=$jobid" /usr/local/logs/swarm_on_slurm.log)
[[ -n $ret ]] || { echo "no swarm found for jobid = $jobid"; exit; }
echo $ret | tr ';' '\n'
$ jobhist 22038972
$ swarmhist 22038972
date=(SKIP)
host=(SKIP)
jobid=22038972
user=(SKIP)
pwd=(SKIP)
ncmds=1
soptions=--array=0-0 --job-name=swarm --output(SKIP)
njobs=1
job-name=swarm
command=/usr/local/bin/swarm -t 16 -g 20 -f outputs/run_seqtools_vc --module samtools,picard --verbose 1
Show how busy is one node: jobload -n cnXXXX
This will show many jobs running in a node. It'll show USER, JOBID, NODECPUS, TOTALCPUS and TOTALNODES (1).
Show properties of a node: freen -n
Use freen -n.
This is helpful if we want to know the node that is allocated from the output (Nodelist column) of the sjobs command.
$ freen -n
........Per-Node Resources........
Partition FreeNds FreeCPUs Cores CPUs Mem Disk Features Nodelist
----------------------------------------------------------------------------------------------------------------------------------
norm* 160/454 17562/25424 28 56 248g 400g cpu56,core28,g256,ssd400,x2695,ibfdr cn[1721-2203,2900-2955]
norm* 0/476 5900/26656 28 56 250g 800g cpu56,core28,g256,ssd800,x2680,ibfdr cn[3092-3631]
norm* 278/309 8928/9888 16 32 123g 800g cpu32,core16,g128,ssd800,x2650,10g cn[0001-0310]
norm* 281/281 4496/4496 8 16 21g 200g cpu16,core8,g24,sata200,x5550,1g cn[2589-2782,2799-2899]
norm* 10/10 160/160 8 16 68g 200g cpu16,core8,g72,sata200,x5550,1g cn[2783-2798]
...
$ sjobs
User JobId JobNa Part St Reason Runtime Walltime Nodes CPUs Memory Dependey Nodelist
=============================================================================================================================
XXX 51944300 sinteracti interactive R 1:13:32 8:00:00 1 16 32 GB cn0862
XXX 51946396 myjob norm R 2:57 12:00:00 1 10 32 GB cn0925
=============================================================================================================================
Exit code
https://hpc.nih.gov/docs/b2-userguide.html#exitcodes
Walltime limits
$ batchlim Max jobs per user: 4000 Max array size: 1001 Partition MaxCPUsPerUser DefWalltime MaxWalltime --------------------------------------------------------------- norm 7360 02:00:00 10-00:00:00 multinode 7560 08:00:00 10-00:00:00 turbo qos 15064 08:00:00 interactive 64 08:00:00 1-12:00:00 (3 simultaneous jobs) quick 6144 02:00:00 04:00:00 largemem 512 02:00:00 10-00:00:00 gpu 728 02:00:00 10-00:00:00 (56 GPUs per user) unlimited 128 UNLIMITED UNLIMITED student 32 02:00:00 08:00:00 (2 GPUs per user) ccr 3072 04:00:00 10-00:00:00 ccrgpu 448 04:00:00 10-00:00:00 (32 GPUs per user) forgo 5760 1-00:00:00 3-00:00:00
Quick jobs
Use the --partition=norm,quick option when you submit a job that requires < 4 hours; see User Guide.
Interactive debugging
Default is 2 CPUs, 4G memory (too small) and 8 hours walltime.
Increase them to 60 GB and more cores if we run something like STAR for rna-seq reads alignment.
sinteractive --mem=32g -c 16 --gres=lscratch:100 --time=24:00:00
The '--gres' option will allocate a local disk, 100GB in this case. The local disk directory will be /lscratch/$SLURM_JOBID.
For RStudio, the example shows to allocate 5GB of temporary space.
For R, it also recommends to allocate a minimal amount of lscratch of 1GB plus whatever lscratch storage is required by your code.
ssh cnXXX
If we have an interactive job on a certain node, we can directly ssh into that node to check for example the /lscratch/$SLURM_JOBID usage. The JobID can be obtained from the jobload command.
It also helps to modify ".ssh/config" to include Host cn* lines. See VS Code on Biowulf and How to Add SSH-Keys to SSH-Agent on Startup in MacOS Operating System.
We can also use freen -n to see some properties of the node.
Parallel jobs
Parallel (MPI) jobs that run on more than 1 node: Use the environment variable $SLURM_NTASKS within the script to specify the number of MPI processes.
Memory and CPUs
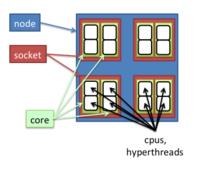
Many nodes has 28 cpus and 256GB memory. The freen command shows 247g for these 256GB node. Luckily in my application, the run time does not change much after I allocate >= 20 CPUs and I don't need that large amount of memory.
Reproduciblity/Pipeline
Singularity
- (obsolete) http://singularity.lbl.gov/, (new) https://sylabs.io/singularity/, (github release) https://github.com/sylabs/singularity,
https://apptainer.org/documentation/ - https://hpc.nih.gov/apps/singularity.html
- .sif (Singularity Image File) is an image file, not a definition file (*.def)
- sif file is created using the sudo singularity build command on a system where you have sudo or root access. Below is an example of my_image.def.
Bootstrap: docker From: ubuntu:20.04 %post apt-get update && apt-get install -y python3- Build the Singularity image (it looks better to use an extension name for easy management later):
sudo singularity build my_image.sif my_image.def sudo singularity build my_image.simg my_image.def sudo singularity build my_image my_image.def
- The definition file is a text file. It does not have to have an extension name. This is like a Dockerfile used by Docker compose.
- Cache images are located in $HOME/.singularity/cache by default. But we can modify it by
export SINGULARITY_CACHEDIR=/data/${USER}/.singularity
- singularity cache list -v cannot return the image names. Singularity equivalent to "docker image list".
- "singularity shell --bind /data/$USER,/fdb,/lscratch my-container.sif" will start a container
- In Singularity, if you don’t specify the destination path in the container when using the --bind option, the destination path will be set equal to the source path. See Bind Paths and Mounts from Singularity User Guide.
- Normally we use the singularity shell or singularity run command to use a singularity image. However, a singularity file can be made so the created image file works like an executable/binary file. See the "rnaseq.def" example in Biowulf. PS: the definition file on Biowulf gave errors. A modified working one is available on Github. Use the bioconda instead of the conda channel. See Installation instructions for RNA-seq analysis using a conda environment V.1.
$ ./rnaseq3 cat /etc/os-release PRETTY_NAME="Debian GNU/Linux 11 (bullseye)" <- Container $ cat /etc/os-release PRETTY_NAME="Ubuntu 22.04.4 LTS" <- Host
- Install from source (in order to obtain a certain version of Singularity). V4.1. It works on Debian 12.
- NCI Containers and Workflows Interest Group
- R and Singularity
- Building a reproducible workflow with Snakemake and Singularity
Definition file
- Definition files from the current version V4.1.
$ singularity exec --app app1 my_container.sif $ singularity exec --app app2 my_container.sif
- Does the definition file depend on Singularity version?
- Yes, the Singularity definition file can depend on the version of Singularity you are using. Different versions of Singularity may support different features, options, and syntax in the definition file.
- For example, some options or features that are available in newer versions of Singularity might not be available in older versions. Conversely, some options or syntax that were used in older versions of Singularity might be deprecated or changed in newer versions.
- Singularity Example Workflow
Import docker images
# Step 1: create a docker archive file docker save -o dockerimage.tar <image_name> # Step 2: convert docker image to singularity image singularity build myimage.sif docker-archive://dockerimage.tar
exec, run, shell
- singularity exec <container> <command>: This command allows you to execute a specific command (required) inside a Singularity container. It starts a container based on the specified image and runs the command provided on the command line following singularity exec <image file name>. This will override any default command specified within the image metadata that would otherwise be run if you used singularity run.
- singularity run: This command runs the %runscript section of the Singularity definition file. If you have specified a command in the %runscript section (like starting the httpgd server), this command will be executed. This is useful when you want to use the Singularity image to perform a specific task or run a specific service.
- singularity shell: This command starts an interactive shell session inside the Singularity container. You can use this command to explore the file system of the Singularity image, install additional packages, or run commands interactively. This is useful for debugging or when you want to interactively work with the software installed in the Singularity image.
Tips:
- it seems "singularity run" and "singularity shell" do not expect any command as an argument.
- if we specify %runscript in a definition, we can use the image by either
- singularity exec XXX.sif /bin/bash # will ignore the %runscript and call the command
- singularity run XXX.sif # will use the %runscript section
- singularity shell XXX.sif # will ignore the %runscript section and launch a shell (/bin/sh)
Bioconductor + httpgd
See Docker -> R and httpgd package.
For some reason, I see the port is open from the netstat command but FF cannot access it. "curl localhost:8888" showed the detail.
Running services
https://docs.sylabs.io/guides/4.1/user-guide/running_services.html
singularity pull library://alpine # Starting Instances singularity instance start alpine_latest.sif instance1 singularity instance list singularity instance start alpine_latest.sif instance2 # Interact with instances singularity exec instance://instance1 cat /etc/os-release singularity run instance://instance2 # run the %runscript for the container singularity shell instance://instance2 # Stopping Instances singularity instance stop instance1 singularity instance stop --all # PS. if an instance was launched by "sudo", we need to use # "sudo" to `list` and `stop` the instance. # “Hello-world” sudo singularity build nginx.sif nginx.def sudo singularity instance start --writable-tmpfs nginx.sif web # INFO: instance started successfully curl localhost sudo singularity instance stop web
Check out the documentation for "API Server Example" where port 9000 was used. In that case, we do not need to use "sudo" to run the service. Below is a working example of running nginx on a non-default port. First, create a file default.conf.
events {
worker_connections 1024;
}
http {
server {
listen 8080;
location / {
root /usr/share/nginx/html;
index index.html index.htm;
}
}
}
Second, create a definition file nginx.def.
Bootstrap: docker
From: nginx
%startscript
nginx -c /etc/nginx/conf.d/default.conf
%files
/home/brb/singularity/nginx/default.conf /etc/nginx/conf.d/default.conf
Last, build the image and run the service.
sudo singularity build nginx.sif nginx.def sudo singularity instance start --writable-tmpfs nginx.sif web2 curl localhost:8080 sudo singularity instance stop -a
Applications
- ZARP-cli & ZARP for Automated RNA-seq Pipeline. See RNA-Seq analysis made easy
Singularity vs Podman
- Singularity vs Podman - Podman and Singularity have different strengths and are suited for different use cases. Podman is more similar to Docker and is suitable for general-purpose containerization, while Singularity is more specialized for use in HPC and scientific computing environments.
- Rootless Containers:
- Podman: Podman allows for running containers as non-root users, which can be useful for security and isolation.
- Singularity: Singularity also supports running containers as non-root users, but it is primarily designed for environments where users do not have root access, such as HPC systems.
- Image Building:
- Podman: Podman has built-in support for building images using a Dockerfile or OCI (Open Container Initiative) image specification.
- Singularity: Singularity uses a different image format and requires a Singularity definition file to build images, which can be more complex than using a Dockerfile.
- Image Distribution:
- Podman: Podman can pull and push images from and to various container registries, similar to Docker.
- Singularity: Singularity images are typically distributed as single files (.sif files) and can be more easily shared and moved between systems.
- User Experience:
- Podman: Podman is designed to have a command-line interface similar to Docker, making it easy for Docker users to transition to Podman.
- Singularity: Singularity has its own set of commands and is designed with a focus on compatibility with existing HPC workflows and environments.
- Use Cases:
- Podman: Podman is suitable for a wide range of use cases, including development, testing, and production environments.
- Singularity: Singularity is more commonly used in HPC and scientific computing environments, where users need to run containers on shared systems without root access.
- Rootless Containers:
- Why Singularity is better than Podman? Singularity has some features that are not available in Podman, which make it particularly well-suited for certain use cases, especially in high-performance computing (HPC) environments:
- Compatibility with HPC Environments: Singularity is designed to work seamlessly in HPC environments where users often lack administrative privileges and need to run containers on shared systems. Its ability to run containers without requiring root access and its support for common HPC features make it a popular choice in these environments.
- Secure Execution: Singularity's security model, which runs containers with the same permissions as the user who launched them, provides a level of security and isolation that is well-suited for environments where security is a concern.
- Image Format: Singularity uses a different image format (.sif files) than Podman (OCI image format). Singularity images are typically distributed as single files, which can be more convenient for sharing and moving between systems, especially in environments where network access is limited.
- A singularity image is portable across platforms, like Snap/Flatpak/AppImage in Linux. For example, if I use "singularity build r-base docker://r-base" to download the latest R image on a Rocky linux (host OS), then the image can be directly run on a Ubuntu linux (host OS).
- Environment Preservation: Singularity is designed to preserve the user's environment inside the container, including environment variables, file system mounts, and user identity. This makes it easier to replicate the user's environment inside the container, which can be important in HPC and scientific computing workflows.
- For the example of an r-base image, an X11 window will open when I draw a plot inside a singularity container.
- Why Podman is better than Singularity? Podman has some features that are not available in Singularity, which make it particularly useful for certain use cases:
- Rootless Containers: Podman allows users to run containers as non-root users, providing a level of security and isolation that is useful in multi-user environments. Singularity also supports running containers as non-root users, but Podman's approach may be more flexible and easier to use in some cases.
- Buildah Integration: Podman integrates with Buildah, a tool for building OCI (Open Container Initiative) container images. This integration allows users to build and manage container images using the same toolset, providing a seamless workflow for container development.
- Docker Compatibility: Podman is designed to be compatible with the Docker CLI (Command Line Interface), making it easy for Docker users to transition to Podman. This compatibility extends to Docker Compose, allowing users to use Podman to manage multi-container applications defined in Docker Compose files.
- Container Management: Podman provides a range of commands for managing containers, including starting, stopping, and removing containers, as well as inspecting container logs and stats. While Singularity also provides similar functionality, Podman's CLI may be more familiar to users accustomed to Docker.
- Networking: Podman provides advanced networking features, including support for CNI (Container Network Interface) plugins, which allow users to configure custom networking solutions for their containers. Singularity's networking capabilities are more limited in comparison.
Snakemake and python
- Snakemake is a workflow management system that aims to reduce the complexity of creating workflows by providing a fast and comfortable execution environment, together with a clean and modern specification language in python style.
- https://snakemake.readthedocs.io/en/stable/
- https://hpc.nih.gov/apps/snakemake.html
- Install on Ubuntu. Before installing Snakemake, you need to
- Install Conda.
- Install Mamba
$ conda install mamba -n base -c conda-forge
- Install snakemake
$ conda activate base $ mamba create -c conda-forge -c bioconda -n snakemake snakemake
- Finally
$ conda activate snakemake $ snakemake --help
Apps
Scientific Applications on NIH HPC Systems
scp
Copy a file from helix to local.
How to handle the path contains spaces.
scp helix:"/data/xxx/ABC abc/file.txt" /tmp/
The double quotes also work for local directories/files.
R program
https://hpc.nih.gov/apps/R.html
Find available R versions:
module -r avail '^R$'
where -r means to use regular expression match. This will match "R/3.5.2" or "R/3.5" but not "Rstudio/1.1.447".
pacman
Biowulf website used the pacman package to manage R packages. Interestingly, pacman is also a command used by Arch Linux.
(Self-installed) R package directory
On our systems, the default path to the library is ~/R/<ver>/library where where ver is the two digit version of R (e.g. 3.5). However, R won't automatically create that directory and in its absence will try to install to the central packge library which will fail. To install packages in your home directory manually create ~/R/<ver>/library first.
The directory ~/R/x86_64-pc-linux-gnu-library/ was not used anymore in Biowulf.
> .libPaths()
[1] "/usr/local/apps/R/4.2/site-library_4.2.0"
[2] "/usr/local/apps/R/4.2/4.2.0/lib64/R/library"
> packageVersion("Greg")
Error in packageVersion("Greg") : there is no package called ‘Greg’
> install.packages("Greg")
Installing package into ‘/usr/local/apps/R/4.2/site-library_4.2.0’
(as ‘lib’ is unspecified)
Warning in install.packages("Greg") :
'lib = "/usr/local/apps/R/4.2/site-library_4.2.0"' is not writable
Would you like to use a personal library instead? (yes/No/cancel) yes
Would you like to create a personal library
‘/home/XXXXXX/R/4.2/library’
to install packages into? (yes/No/cancel) yes
--- Please select a CRAN mirror for use in this session ---
Secure CRAN mirrors
1: 0-Cloud [https]
...
74: USA (IA) [https]
75: USA (MI) [https]
76: USA (OH) [https]
77: USA (OR) [https]
78: USA (TN) [https]
79: USA (TX 1) [https]
80: Uruguay [https]
81: (other mirrors)
Selection: 74
trying URL 'https://mirror.las.iastate.edu/CRAN/src/contrib/Greg_1.4.1.tar.gz'
Content type 'application/x-gzip' length 205432 bytes (200 KB)
> .libPaths()
[1] "/spin1/home/linux/XXXXXX/R/4.2/library"
[2] "/usr/local/apps/R/4.2/site-library_4.2.0"
[3] "/usr/local/apps/R/4.2/4.2.0/lib64/R/library"
RStudio (standalone application)
This approach requires running the NoMachine app.
- Step 1: Connecting to Biowulf with NoMachine (NX) (Tip: no need to use ssh from a terminal)
- It works though the resolution is not great on a Mac screen. This assume we are in the NIH network.
- One thing is about adjusting the window size. Click Ctrl + Alt + 0 to bring up the setting if we accept all the default options when we connect to a remote machine. Click 'Resize remote display'. Now we can use mouse to drag and adjust the display size. This will keep the resolution as it showed up originally. It is easier when I choose the 'key-based authorization method with a key you provide'.
- Desktop environment is XFCE ($DESKTOP_SESSION)
- Step 2: Use RStudio on Biowulf.
- We need to open a terminal app from the RedHat OS desktop and follow the instruction there.
- The desktop IDE (NOT a web application) program is installed in /usr/local/apps/rstudio/rstudio-1.4.1103/.
- Problems: The copy/paste keyboard shortcuts do not work even I checked the option "Grab the keyboard input". Current resolution: Use two ssh connections (one to run R, and another one to edit file). Open noMachine and use the desktop to view graph files (gv XXX.pdf OR change the default program to 'GNU GV Postscript/PDF Viewer' from the default 'LibreOffice Draw').
sinteractive --mem=6g --gres=lscratch:5 module load Rstudio R rstudio &
httpgd: Web application
This approach just need a Terminal and a browser. SSH tunnel is used. The port number is shown in the output of "sinteractive --tunnel" command.
Refer to the instruction for the Jupyter case. The key is the port number has to be passed to R when we call hgd() function.
- Terminal 1
ssh biowulf.nih.gov sinteractive --gres=lscratch:5 --mem=10g --tunnel # Please create a SSH tunnel from your workstation to these ports on biowulf. # On Linux/MacOS, open a terminal and run: # # ssh -L 33327:localhost:33327 [email protected] module load R/4.3 R > library(httpgd) > httpgd::hgd(port = as.integer(Sys.getenv("PORT1"))) httpgd server running at: http://127.0.0.1:33327/live?token=FpaxiBKz
- Terminal 2
ssh -L 33327:localhost:33327 biowulf.nih.gov
Open the URL http://127.0.0.1:33327/live?token=FpaxiBKz given by httpgd::hgd() on my local browser.
Call "exit" twice on Terminal 1 and "exit" once on Terminal 2 to quit Biowulf.
Python and conda
- https://hpc.nih.gov/apps/python.html. Search pip.
- Jupyter on Biowulf
Visual Studio Code
- VS Code on Biowulf. Install the "Remote Development" VScode extension.
- Run Jupyter notebook on a compute node in VS Code.
- Remote Development using SSH. Basically once we connect to a remote host, a new VScode instance will be created and we can open any files from the remote host on VScode.
Exit an SSH session
Today after I issued a "cd /data/USERNAME" command, it just hung there even the Ctrl+c won't exit.
One solution is to close the current terminal. If we like to keep the current terminal, a solution is to open another terminal, give an SSH connection and run pkill -u USERNAME. This will also kill all the SSH connections including the current one.
SSH tunnel
https://hpc.nih.gov/docs/tunneling/
The use of interactive application servers (such as Jupyter notebooks) on Biowulf compute nodes requires establishing SSH tunnels to make the service accessible to your local workstation.
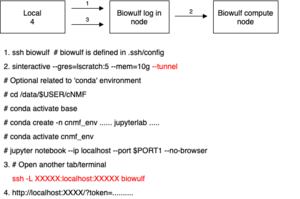
Jupyter
The following diagram contains steps to launch Jupyter with an optional conda environment.
Terminal customization
- ssh add authorized_keys
- ~/.bashrc:
- change PS1
- add an alias for nano
- ~/.bash_profile: no change
- ~/.nanorc, ~/r.nanorc and ~/bin/nano/bin/nano (4.2)
- ~/.emacs: global-display-line-numbers-mode
- ~/.vimrc: set number
- .Rprofile: options(editor="emacs")
- .bash_logout: no change
tmux for keeping SSH sessions
Reference genomes
$ wc -l /fdb/STAR/iGenomes/Homo_sapiens/UCSC/hg19/genes.gtf 1006819 /fdb/STAR/iGenomes/Homo_sapiens/UCSC/hg19/genes.gtf $ wc -l /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf 1006819 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf # Transcript $ grep uc002gig /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/* /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/kgXref.txt:uc002gig.1 AM076971 E7EQX7 E7EQX7_HUMAN TP53 NM_001126114 NP_001119586 Homo sapiens tumor protein p53 (TP53), transcript variant 2, mRNA. /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownGene.txt:uc002gig.1 chr17 - 7565096 7579937 75652567579912 7 7565096,7577498,7578176,7578370,7579311,7579699,7579838, 7565332,7577608,7578289,7578554,7579590,7579721,7579937, E7EQX7 uc002gig.1 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownToRefSeq.txt:uc002gig.1 NM_001126114 $ ls -logh /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/ total 231M -rwxrwxr-x 1 31K Jul 21 2015 cytoBand.txt -rwxrwxr-x 1 136M Jul 21 2015 genes.gtf -rwxrwxr-x 1 11M Jul 21 2015 kgXref.txt -rwxrwxr-x 1 20M Jul 21 2015 knownGene.txt -rwxrwxr-x 1 1.6M Jul 21 2015 knownToRefSeq.txt -rwxrwxr-x 1 4.6M Jul 21 2015 refFlat.txt.gz -rwxrwxr-x 1 15M Jul 26 2015 refGene.txt -rwxrwxr-x 1 45M Jul 21 2015 refSeqSummary.txt $ wc -l /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownGene.txt 82960 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownGene.txt $ wc -l /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf 1006819 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf $ head -4 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/genes.gtf chr1 unknown exon 11874 12227 . + . gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS16932"; chr1 unknown exon 12613 12721 . + . gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS16932"; chr1 unknown exon 13221 14409 . + . gene_id "DDX11L1"; gene_name "DDX11L1"; transcript_id "NR_046018"; tss_id "TSS16932"; chr1 unknown exon 14362 14829 . - . gene_id "WASH7P"; gene_name "WASH7P"; transcript_id "NR_024540"; tss_id "TSS8568"; $ head -4 /fdb/igenomes/Homo_sapiens/UCSC/hg19/Annotation/Genes/knownGene.txt uc001aaa.3 chr1 + 11873 14409 11873 11873 3 11873,12612,13220, 12227,12721,14409, uc001aaa.3 uc010nxr.1 chr1 + 11873 14409 11873 11873 3 11873,12645,13220, 12227,12697,14409, uc010nxr.1 uc010nxq.1 chr1 + 11873 14409 12189 13639 3 11873,12594,13402, 12227,12721,14409, B7ZGX9 uc010nxq.1 uc009vis.3 chr1 - 14361 16765 14361 14361 4 14361,14969,15795,16606, 14829,15038,15942,16765, uc009vis.3