R: Difference between revisions
| Line 371: | Line 371: | ||
* https://www.rstudio.com/resources/videos/plumber-turning-your-r-code-into-an-api/ | * https://www.rstudio.com/resources/videos/plumber-turning-your-r-code-into-an-api/ | ||
* [https://blog.rstudio.com/2018/10/23/rstudio-1-2-preview-plumber-integration/ RStudio 1.2 Preview: Plumber Integration] | * [https://blog.rstudio.com/2018/10/23/rstudio-1-2-preview-plumber-integration/ RStudio 1.2 Preview: Plumber Integration] | ||
* [https://medium.com/@skyetetra/using-docker-to-deploy-an-r-plumber-api-863ccf91516d Using docker to deploy an R plumber API] | |||
=== Docker === | === Docker === | ||
Revision as of 14:41, 2 January 2019
Install and upgrade R
Online Editor
We can run R on web browsers without installing it on local machines (similar to [/ideone.com Ideone.com] for C++. It does not require an account either (cf RStudio).
rdrr.io
It can produce graphics too. The package I am testing (cobs) is available too.
rstudio.cloud
RDocumentation
The interactive engine is based on DataCamp Light
For example, tbl_df function from dplyr package.
The website DataCamp allows to run library() on the Script window. After that, we can use the packages on R Console.
Here is a list of (common) R packages that users can use on the web.
The packages on RDocumentation may be outdated. For example, the current stringr on CRAN is v1.2.0 (2/18/2017) but RDocumentation has v1.1.0 (8/19/2016).
Web Applications
See also CRAN Task View: Web Technologies and Services
TexLive
TexLive can be installed by 2 ways
- Ubuntu repository; does not include tlmgr utility for package manager.
- Official website
texlive-latex-extra
https://packages.debian.org/sid/texlive-latex-extra
For example, framed and titling packages are included.
tlmgr - TeX Live package manager
https://www.tug.org/texlive/tlmgr.html
TinyTex
https://github.com/yihui/tinytex
pkgdown: create a website for your package
Building a website with pkgdown: a short guide
Rmarkdown: create HTML5 web, slides and more
- http://rmarkdown.rstudio.com/html_document_format.html
- R Markdown: Eight ways
- https://www.rstudio.com/wp-content/uploads/2015/02/rmarkdown-cheatsheet.pdf
- https://www.rstudio.com/wp-content/uploads/2015/03/rmarkdown-reference.pdf
- Chunk options http://kbroman.org/knitr_knutshell/pages/Rmarkdown.html
HTML5 slides examples
- http://yihui.name/slides/knitr-slides.html
- http://yihui.name/slides/2012-knitr-RStudio.html
- http://yihui.name/slides/2011-r-dev-lessons.html#slide1
- http://inundata.org/R_talks/BARUG/#intro
Software requirement
- Rstudio
- knitr, XML, RCurl (See omegahat or this internal link for installation on Ubuntu)
- pandoc package This is a command line tool. I am testing it on Windows 7.
Slide #22 gives an instruction to create
- regular html file by using RStudio -> Knit HTML button
- HTML5 slides by using pandoc from command line.
Files:
- Rcmd source: 009-slides.Rmd Note that IE 8 was not supported by github. For IE 9, be sure to turn off "Compatibility View".
- markdown output: 009-slides.md
- HTML output: 009-slides.html
We can create Rcmd source in Rstudio by File -> New -> R Markdown.
There are 4 ways to produce slides with pandoc
- S5
- DZSlides
- Slidy
- Slideous
Use the markdown file (md) and convert it with pandoc
pandoc -s -S -i -t dzslides --mathjax html5_slides.md -o html5_slides.html
If we are comfortable with HTML and CSS code, open the html file (generated by pandoc) and modify the CSS style at will.
Built-in examples from rmarkdown
# This is done on my ODroid xu4 running Ubuntu Mate 15.10 (Wily)
# I used sudo apt-get install pandoc in shell
# and install.packages("rmarkdown") in R 3.2.3
library(rmarkdown)
rmarkdown::render("~/R/armv7l-unknown-linux-gnueabihf-library/3.2/rmarkdown/rmarkdown/templates/html_vignette/skeleton/skeleton.Rmd")
# the output <skeleton.html> is located under the same dir as <skeleton.Rmd>
Note that the image files in the html are embedded Base64 images in the html file. See
- http://stackoverflow.com/questions/1207190/embedding-base64-images
- Data URI scheme
- http://www.r-bloggers.com/embed-images-in-rd-documents/
- How to not embed Base64 images in RMarkdown
- Upload plots as PNG file to your wordpress
Templates
- https://github.com/rstudio/rticles/tree/master/inst/rmarkdown/templates
- https://github.com/rstudio/rticles/blob/master/inst/rmarkdown/templates/jss_article/resources/template.tex
Knit button
- It calls rmarkdown::render()
- R Markdown = knitr + Pandoc
- rmarkdown::render () = knitr::knit() + a system() call to pandoc
Pandoc's Markdown
Originally Pandoc is for html.
Extensions
- YAML metadata
- Latex Math
- syntax highlight
- embed raw HTML/Latex (raw HTML only works for HTML output and raw Latex only for Latex/pdf output)
- tables
- footnotes
- citations
Types of output documents
- Latex/pdf, HTML, Word
- beamer, ioslides, Slidy, reval.js
- Ebooks
- ...
Some examples:
pandoc test.md -o test.html pandoc test.md -s --mathjax -o test.html pandoc test.md -o test.docx pandoc test.md -o test.pdf pandoc test.md --latex-engine=xlelatex -o test.pdf pandoc test.md -o test.epb
Check out ?rmarkdown::pandoc_convert()/
When you click the Knit button in RStudio, you will see the actual command that is executed.
Global options
Suppose I want to create a simple markdown only documentation without worrying about executing code, instead of adding eval = FALSE to each code chunks, I can insert the following between YAML header and the content. Even bash chunks will not be executed.
```{r setup, include=FALSE}
knitr::opts_chunk$set(echo = TRUE, eval = FALSE)
```
Examples/gallery
Some examples of creating papers (with references) based on knitr can be found on the Papers and reports section of the knitr website.
- https://rmarkdown.rstudio.com/gallery.html
- https://github.com/EBI-predocs/knitr-example
- https://github.com/timchurches/meta-analyses
- http://www.gastonsanchez.com/depot/knitr-slides
Read the docs Sphinx theme and journal article formats
http://blog.rstudio.org/2016/03/21/r-markdown-custom-formats/
- rticles package
- rmdformats package
rmarkdown news
Useful tricks when including images in Rmarkdown documents
http://blog.revolutionanalytics.com/2017/06/rmarkdown-tricks.html
Converting Rmarkdown to F1000Research LaTeX Format
BiocWorkflowTools package and paper
icons for rmarkdown
https://ropensci.org/technotes/2018/05/15/icon/
Reproducible data analysis
Automatic document production with R
https://itsalocke.com/improving-automatic-document-production-with-r/
Documents with logos, watermarks, and corporate styles
http://ellisp.github.io/blog/2017/09/09/rmarkdown
rticles and pinp for articles
- https://cran.r-project.org/web/packages/rticles/index.html
- http://dirk.eddelbuettel.com/code/pinp.html
Gmisc: create Table 1 used in medical articles
https://cran.r-project.org/web/packages/Gmisc/index.html
Markdown language
According to wikipedia:
Markdown is a lightweight markup language, originally created by John Gruber with substantial contributions from Aaron Swartz, allowing people “to write using an easy-to-read, easy-to-write plain text format, then convert it to structurally valid XHTML (or HTML)”.
- Markup is a general term for content formatting - such as HTML - but markdown is a library that generates HTML markup.
- Nice summary from stackoverflow.com and more complete list from github.
- An example https://gist.github.com/jeromyanglim/2716336
- Convert mediawiki to markdown using online conversion tool from pandoc.
- R markdown file and use it in RStudio. Customizing Chunk Options can be found in knitr page and rpubs.com.
RStudio
RStudio is the best editor.
Markdown has two drawbacks: 1. it does not support TOC natively. 2. RStudio cannot show headers in the editor.
Therefore, use rmarkdown format instead of markdown.
Blogdown
- https://github.com/rstudio/blogdown
- https://bookdown.org/yihui/blogdown/
- BLOGDOWN TUTORIAL Peter's blog
- Customizing Hugo / Blogdown RSS Templates
HTTP protocol
- http://en.wikipedia.org/wiki/File:Http_request_telnet_ubuntu.png
- Query string
- How to capture http header? Use curl -i en.wikipedia.org.
- Web Inspector. Build-in in Chrome. Right click on any page and choose 'Inspect Element'.
- Web server
- Simple TCP/IP web server
- HTTP Made Really Easy
- Illustrated Guide to HTTP
- nweb: a tiny, safe Web server with 200 lines
- Tiny HTTPd
An HTTP server is conceptually simple:
- Open port 80 for listening
- When contact is made, gather a little information (get mainly - you can ignore the rest for now)
- Translate the request into a file request
- Open the file and spit it back at the client
It gets more difficult depending on how much of HTTP you want to support - POST is a little more complicated, scripts, handling multiple requests, etc.
Example in R
> co <- socketConnection(port=8080, server=TRUE, blocking=TRUE) > # Now open a web browser and type http://localhost:8080/index.html > readLines(co,1) [1] "GET /index.html HTTP/1.1" > readLines(co,1) [1] "Host: localhost:8080" > readLines(co,1) [1] "User-Agent: Mozilla/5.0 (X11; Ubuntu; Linux i686; rv:23.0) Gecko/20100101 Firefox/23.0" > readLines(co,1) [1] "Accept: text/html,application/xhtml+xml,application/xml;q=0.9,*/*;q=0.8" > readLines(co,1) [1] "Accept-Language: en-US,en;q=0.5" > readLines(co,1) [1] "Accept-Encoding: gzip, deflate" > readLines(co,1) [1] "Connection: keep-alive" > readLines(co,1) [1] ""
Example in C (Very simple http server written in C, 187 lines)
Create a simple hello world html page and save it as <index.html> in the current directory (/home/brb/Downloads/)
Launch the server program (assume we have done gcc http_server.c -o http_server)
$ ./http_server -p 50002 Server started at port no. 50002 with root directory as /home/brb/Downloads
Secondly open a browser and type http://localhost:50002/index.html. The server will respond
GET /index.html HTTP/1.1 Host: localhost:50002 User-Agent: Mozilla/5.0 (X11; Ubuntu; Linux i686; rv:23.0) Gecko/20100101 Firefox/23.0 Accept: text/html,application/xhtml+xml,application/xml;q=0.9,*/*;q=0.8 Accept-Language: en-US,en;q=0.5 Accept-Encoding: gzip, deflate Connection: keep-alive file: /home/brb/Downloads/index.html GET /favicon.ico HTTP/1.1 Host: localhost:50002 User-Agent: Mozilla/5.0 (X11; Ubuntu; Linux i686; rv:23.0) Gecko/20100101 Firefox/23.0 Accept: text/html,application/xhtml+xml,application/xml;q=0.9,*/*;q=0.8 Accept-Language: en-US,en;q=0.5 Accept-Encoding: gzip, deflate Connection: keep-alive file: /home/brb/Downloads/favicon.ico GET /favicon.ico HTTP/1.1 Host: localhost:50003 User-Agent: Mozilla/5.0 (X11; Ubuntu; Linux i686; rv:23.0) Gecko/20100101 Firefox/23.0 Accept: text/html,application/xhtml+xml,application/xml;q=0.9,*/*;q=0.8 Accept-Language: en-US,en;q=0.5 Accept-Encoding: gzip, deflate Connection: keep-alive file: /home/brb/Downloads/favicon.ico
The browser will show the page from <index.html> in server.
The only bad thing is the code does not close the port. For example, if I have use Ctrl+C to close the program and try to re-launch with the same port, it will complain socket() or bind(): Address already in use.
Another Example in C (55 lines)
http://mwaidyanatha.blogspot.com/2011/05/writing-simple-web-server-in-c.html
The response is embedded in the C code.
If we test the server program by opening a browser and type "http://localhost:15000/", the server received the follwing 7 lines
GET / HTTP/1.1 Host: localhost:15000 User-Agent: Mozilla/5.0 (X11; Ubuntu; Linux i686; rv:23.0) Gecko/20100101 Firefox/23.0 Accept: text/html,application/xhtml+xml,application/xml;q=0.9,*/*;q=0.8 Accept-Language: en-US,en;q=0.5 Accept-Encoding: gzip, deflate Connection: keep-alive
If we include a non-executable file's name in the url, we will be able to download that file. Try "http://localhost:15000/client.c".
If we use telnet program to test, wee need to type anything we want
$ telnet localhost 15000 Trying 127.0.0.1... Connected to localhost. Escape character is '^]'. ThisCanBeAnything <=== This is what I typed in the client and it is also shown on server HTTP/1.1 200 OK <=== From here is what I got from server Content-length: 37Content-Type: text/html HTML_DATA_HERE_AS_YOU_MENTIONED_ABOVE <=== The html tags are not passed from server, interesting! Connection closed by foreign host. $
See also more examples under C page.
Others
- http://rosettacode.org/wiki/Hello_world/ (Different languages)
- http://kperisetla.blogspot.com/2012/07/simple-http-web-server-in-c.html (Windows web server)
- http://css.dzone.com/articles/web-server-c (handling HTTP GET request, handling content types(txt, html, jpg, zip. rar, pdf, php etc.), sending proper HTTP error codes, serving the files from a web root, change in web root in a config file, zero copy optimization using sendfile method and php file handling.)
- https://github.com/gtungatkar/Simple-HTTP-server
- https://github.com/davidmoreno/onion
shiny
See Shiny.
plumber: Turning your R code into a RESTful Web API
- https://github.com/trestletech/plumber
- https://www.rstudio.com/resources/videos/plumber-turning-your-r-code-into-an-api/
- RStudio 1.2 Preview: Plumber Integration
- Using docker to deploy an R plumber API
Docker
- Using Docker as a Personal Productivity Tool – Running Command Line Apps Bundled in Docker Containers
- Dockerized RStudio server from Duke University. 110 containers were set up on a cloud server (4 cores, 28GB RAM, 400GB disk). Each container has its own port number. Each student is mapped to a single container. https://github.com/mccahill/docker-rstudio
- RStudio in the cloud with Amazon Lightsail and docker
- Mark McCahill (RStudio + Docker)
- BiocImageBuilder
- Why Use Docker with R? A DevOps Perspective
httpuv
http and WebSocket library.
See also the servr package which can start an HTTP server in R to serve static files, or dynamic documents that can be converted to HTML files (e.g., R Markdown) under a given directory.
RApache
gWidgetsWWW
- http://www.jstatsoft.org/v49/i10/paper
- gWidgetsWWW2 gWidgetsWWW based on Rook
- Compare shiny with gWidgetsWWW2.rapache
Rook
Since R 2.13, the internal web server was exposed.
Tutorual from useR2012 and Jeffrey Horner
Here is another one from http://www.rinfinance.com.
Rook is also supported by [rApache too. See http://rapache.net/manual.html.
Google group. https://groups.google.com/forum/?fromgroups#!forum/rrook
Advantage
- the web applications are created on desktop, whether it is Windows, Mac or Linux.
- No Apache is needed.
- create multiple applications at the same time. This complements the limit of rApache.
4 lines of code example.
library(Rook)
s <- Rhttpd$new()
s$start(quiet=TRUE)
s$print()
s$browse(1) # OR s$browse("RookTest")
Notice that after s$browse() command, the cursor will return to R because the command just a shortcut to open the web page http://127.0.0.1:10215/custom/RookTest.
We can add Rook application to the server; see ?Rhttpd.
s$add(
app=system.file('exampleApps/helloworld.R',package='Rook'),name='hello'
)
s$add(
app=system.file('exampleApps/helloworldref.R',package='Rook'),name='helloref'
)
s$add(
app=system.file('exampleApps/summary.R',package='Rook'),name='summary'
)
s$print()
#Server started on 127.0.0.1:10221
#[1] RookTest http://127.0.0.1:10221/custom/RookTest
#[2] helloref http://127.0.0.1:10221/custom/helloref
#[3] summary http://127.0.0.1:10221/custom/summary
#[4] hello http://127.0.0.1:10221/custom/hello
# Stops the server but doesn't uninstall the app
## Not run:
s$stop()
## End(Not run)
s$remove(all=TRUE)
rm(s)
For example, the interface and the source code of summary app are given below
app <- function(env) {
req <- Rook::Request$new(env)
res <- Rook::Response$new()
res$write('Choose a CSV file:\n')
res$write('<form method="POST" enctype="multipart/form-data">\n')
res$write('<input type="file" name="data">\n')
res$write('<input type="submit" name="Upload">\n</form>\n<br>')
if (!is.null(req$POST())){
data <- req$POST()[['data']]
res$write("<h3>Summary of Data</h3>");
res$write("<pre>")
res$write(paste(capture.output(summary(read.csv(data$tempfile,stringsAsFactors=FALSE)),file=NULL),collapse='\n'))
res$write("</pre>")
res$write("<h3>First few lines (head())</h3>");
res$write("<pre>")
res$write(paste(capture.output(head(read.csv(data$tempfile,stringsAsFactors=FALSE)),file=NULL),collapse='\n'))
res$write("</pre>")
}
res$finish()
}
More example:
- http://lamages.blogspot.com/2012/08/rook-rocks-example-with-googlevis.html
- Self-organizing map
- Deploy Rook apps with rApache. First one and two.
- A Simple Prediction Web Service Using the New fiery Package
sumo
Sumo is a fully-functional web application template that exposes an authenticated user's R session within java server pages. See the paper http://journal.r-project.org/archive/2012-1/RJournal_2012-1_Bergsma+Smith.pdf.
Stockplot
FastRWeb
http://cran.r-project.org/web/packages/FastRWeb/index.html
Rwui
CGHWithR and WebDevelopR
CGHwithR is still working with old version of R although it is removed from CRAN. Its successor is WebDevelopR. Its The vignette (year 2013) provides a review of several available methods.
manipulate from RStudio
This is not a web application. But the manipulate package can be used to create interactive plot within R(Studio) environment easily. Its source is available at here.
Mathematica also has manipulate function for plotting; see here.
RCloud
RCloud is an environment for collaboratively creating and sharing data analysis scripts. RCloud lets you mix analysis code in R, HTML5, Markdown, Python, and others. Much like Sage, iPython notebooks and Mathematica, RCloud provides a notebook interface that lets you easily record a session and annotate it with text, equations, and supporting images.
See also the Talk in UseR 2014.
cloudyr and flyio - Input Output Files in R from Cloud or Local
https://blog.socialcops.com/inside-sc/announcements/flyio-r-package-interact-data-cloud/ Announcing flyio, an R Package to Interact with Data in the Cloud]
Dropbox access
rdrop2 package
Web page scraping
http://www.slideshare.net/schamber/web-data-from-r#btnNext
xml2 package
rvest package depends on xml2.
purrr
- https://purrr.tidyverse.org/
- Getting started with the purrr package in R, especially the map() function.
rvest
On Ubuntu, we need to install two packages first!
sudo apt-get install libcurl4-openssl-dev # OR libcurl4-gnutls-dev sudo apt-get install libxml2-dev
- https://github.com/hadley/rvest
- Visualizing obesity across United States by using data from Wikipedia
- rvest tutorial: scraping the web using R
- https://renkun.me/pipeR-tutorial/Examples/rvest.html
- http://zevross.com/blog/2015/05/19/scrape-website-data-with-the-new-r-package-rvest/
- Google scholar scraping with rvest package
Animate
- Animating Changes in Football Kits using R: rvest, tidyverse, xml2, purrr & magick
- Animated Directional Chord Diagrams tweenr & magick
V8: Embedded JavaScript Engine for R
R⁶ — General (Attys) Distributions: V8, rvest, ggbeeswarm, hrbrthemes and tidyverse packages are used.
pubmed.mineR
Text mining of PubMed Abstracts (http://www.ncbi.nlm.nih.gov/pubmed). The algorithms are designed for two formats (text and XML) from PubMed.
R code for scraping the P-values from pubmed, calculating the Science-wise False Discovery Rate, et al (Jeff Leek)
These R packages import sports, weather, stock data and more
Diving Into Dynamic Website Content with splashr
https://rud.is/b/2017/02/09/diving-into-dynamic-website-content-with-splashr/
Send email
mailR
Easiest. Require rJava package (not trivial to install, see rJava). mailR is an interface to Apache Commons Email to send emails from within R. See also send bulk email
Before we use the mailR package, we have followed here to have Allow less secure apps: 'ON' ; or you might get an error Error: EmailException (Java): Sending the email to the following server failed : smtp.gmail.com:465. Once we turn on this option, we may get an email for the notification of this change. Note that the recipient can be other than a gmail.
> send.mail(from = "[email protected]", to = c("[email protected]", "Recipient 2 <[email protected]>"), replyTo = c("Reply to someone else <[email protected]>") subject = "Subject of the email", body = "Body of the email", smtp = list(host.name = "smtp.gmail.com", port = 465, user.name = "gmail_username", passwd = "password", ssl = TRUE), authenticate = TRUE, send = TRUE) [1] "Java-Object{org.apache.commons.mail.SimpleEmail@7791a895}"
gmailr
More complicated. gmailr provides access the Google's gmail.com RESTful API. Vignette and an example on here. Note that it does not use a password; it uses a json file for oauth authentication downloaded from https://console.cloud.google.com/. See also https://github.com/jimhester/gmailr/issues/1.
library(gmailr)
gmail_auth('mysecret.json', scope = 'compose')
test_email <- mime() %>%
to("[email protected]") %>%
from("[email protected]") %>%
subject("This is a subject") %>%
html_body("<html><body>I wish <b>this</b> was bold</body></html>")
send_message(test_email)
sendmailR
sendmailR provides a simple SMTP client. It is not clear how to use the package (i.e. where to enter the password).
GEO (Gene Expression Omnibus)
See this internal link.
Interactive html output
sendplot
RIGHT
The supported plot types include scatterplot, barplot, box plot, line plot and pie plot.
In addition to tooltip boxes, the package can create a table showing all information about selected nodes.
d3Network
- http://christophergandrud.github.io/d3Network/ (old)
- https://christophergandrud.github.io/networkD3/ (new)
library(d3Network)
Source <- c("A", "A", "A", "A", "B", "B", "C", "C", "D")
Target <- c("B", "C", "D", "J", "E", "F", "G", "H", "I")
NetworkData <- data.frame(Source, Target)
d3SimpleNetwork(NetworkData, height = 800, width = 1024, file="tmp.html")
htmlwidgets for R
Embed widgets in R Markdown documents and Shiny web applications.
- Official website http://www.htmlwidgets.org/.
- How to write a useful htmlwidgets in R: tips and walk-through a real example
networkD3
This is a port of Christopher Gandrud's d3Network package to the htmlwidgets framework.
scatterD3
scatterD3 is an HTML R widget for interactive scatter plots visualization. It is based on the htmlwidgets R package and on the d3.js javascript library.
rthreejs - Create interactive 3D scatter plots, network plots, and globes
d3heatmap
See R
svgPanZoom
This 'htmlwidget' provides pan and zoom interactivity to R graphics, including 'base', 'lattice', and 'ggplot2'. The interactivity is provided through the 'svg-pan-zoom.js' library.
DT: An R interface to the DataTables library
plotly
- Power curves and ggplot2.
- TIME SERIES CHARTS BY THE ECONOMIST IN R USING PLOTLY & FIVE INTERACTIVE R VISUALIZATIONS WITH D3, GGPLOT2, & RSTUDIO
- Filled chord diagram
- DASHBOARDS IN R WITH SHINY & PLOTLY
- Plotly Graphs in Shiny,
- How to plot basic charts with plotly
- How to add Trend Lines in R Using Plotly
Amazon
Download product information and reviews from Amazon.com
sudo apt-get install libxml2-dev sudo apt-get install libcurl4-openssl-dev
and in R
install.packages("devtools")
install.packages("XML")
install.packages("pbapply")
install.packages("dplyr")
devtools::install_github("56north/Rmazon")
product_info <- Rmazon::get_product_info("1593273843")
reviews <- Rmazon::get_reviews("1593273843")
reviews[1,6] # only show partial characters from the 1st review
nchar(reviews[1,6])
as.character(reviews[1,6]) # show the complete text from the 1st review
reviews <- Rmazon::get_reviews("B07BNGJXGS")
# Fetching 30 reviews of 'BOOX Note Ereader,Android 6.0 32 GB 10.3" Dual Touch HD Display'
# |++++++++++++++++++++++++++++++++++++++++++++++++++| 100% elapsed = 02s
reviews
# A tibble: 30 x 6
reviewRating reviewDate reviewFormat Verified_Purcha… reviewHeadline
<dbl> <chr> <lgl> <lgl> <chr>
1 4 May 23, 2… NA TRUE Good for PDF …
2 3 May 8, 20… NA FALSE The reading s…
3 5 May 17, 2… NA TRUE E-reader and …
4 3 May 24, 2… NA TRUE Good hardware…
5 3 June 21, … NA TRUE Poor QC
6 5 August 5,… NA TRUE Excellent for…
7 5 May 31, 2… NA TRUE Especially li…
8 5 July 4, 2… NA TRUE Android 6 rea…
9 4 July 15, … NA TRUE Remember the …
10 4 June 9, 2… NA TRUE Overall fanta…
# ... with 20 more rows, and 1 more variable: reviewText <chr>
reviews[1, 6] # 6-th column is the review text
gutenbergr
OCR
- Tesseract package: High Quality OCR in R, How to do Optical Character Recognition (OCR) of non-English documents in R using Tesseract?
- https://cran.r-project.org/web/packages/abbyyR/index.html
Creating local repository for CRAN and Bioconductor (focus on Windows binary packages only)
How to set up a local repository
- CRAN specific: http://cran.r-project.org/mirror-howto.html
- Bioconductor specific: http://www.bioconductor.org/about/mirrors/mirror-how-to/
- How to Set Up a Custom CRAN-like Repository
General guide: http://cran.r-project.org/doc/manuals/R-admin.html#Setting-up-a-package-repository
Utilities such as install.packages can be pointed at any CRAN-style repository, and R users may want to set up their own. The ‘base’ of a repository is a URL such as http://www.omegahat.org/R/: this must be an URL scheme that download.packages supports (which also includes ‘ftp://’ and ‘file://’, but not on most systems ‘https://’). Under that base URL there should be directory trees for one or more of the following types of package distributions:
- "source": located at src/contrib and containing .tar.gz files. Other forms of compression can be used, e.g. .tar.bz2 or .tar.xz files.
- "win.binary": located at bin/windows/contrib/x.y for R versions x.y.z and containing .zip files for Windows.
- "mac.binary.leopard": located at bin/macosx/leopard/contrib/x.y for R versions x.y.z and containing .tgz files.
Each terminal directory must also contain a PACKAGES file. This can be a concatenation of the DESCRIPTION files of the packages separated by blank lines, but only a few of the fields are needed. The simplest way to set up such a file is to use function write_PACKAGES in the tools package, and its help explains which fields are needed. Optionally there can also be a PACKAGES.gz file, a gzip-compressed version of PACKAGES—as this will be downloaded in preference to PACKAGES it should be included for large repositories. (If you have a mis-configured server that does not report correctly non-existent files you will need PACKAGES.gz.)
To add your repository to the list offered by setRepositories(), see the help file for that function.
A repository can contain subdirectories, when the descriptions in the PACKAGES file of packages in subdirectories must include a line of the form
Path: path/to/subdirectory
—once again write_PACKAGES is the simplest way to set this up.
Space requirement if we want to mirror WHOLE repository
- Whole CRAN takes about 92GB (rsync -avn cran.r-project.org::CRAN > ~/Downloads/cran).
- Bioconductor is big (> 64G for BioC 2.11). Please check the size of what will be transferred with e.g. (rsync -avn bioconductor.org::2.11 > ~/Downloads/bioc) and make sure you have enough room on your local disk before you start.
On the other hand, we if only care about Windows binary part, the space requirement is largely reduced.
- CRAN: 2.7GB
- Bioconductor: 28GB.
Misc notes
- If the binary package was built on R 2.15.1, then it cannot be installed on R 2.15.2. But vice is OK.
- Remember to issue "--delete" option in rsync, otherwise old version of package will be installed.
- The repository still need src directory. If it is missing, we will get an error
Warning: unable to access index for repository http://arraytools.no-ip.org/CRAN/src/contrib Warning message: package ‘glmnet’ is not available (for R version 2.15.2)
The error was given by available.packages() function.
To bypass the requirement of src directory, I can use
install.packages("glmnet", contriburl = contrib.url(getOption('repos'), "win.binary"))
but there may be a problem when we use biocLite() command.
I find a workaround. Since the error comes from missing CRAN/src directory, we just need to make sure the directory CRAN/src/contrib exists AND either CRAN/src/contrib/PACKAGES or CRAN/src/contrib/PACKAGES.gz exists.
To create CRAN repository
Before creating a local repository please give a dry run first. You don't want to be surprised how long will it take to mirror a directory.
Dry run (-n option). Pipe out the process to a text file for an examination.
rsync -avn cran.r-project.org::CRAN > crandryrun.txt
To mirror only partial repository, it is necessary to create directories before running rsync command.
cd mkdir -p ~/Rmirror/CRAN/bin/windows/contrib/2.15 rsync -rtlzv --delete cran.r-project.org::CRAN/bin/windows/contrib/2.15/ ~/Rmirror/CRAN/bin/windows/contrib/2.15 (one line with space before ~/Rmirror) # src directory is very large (~27GB) since it contains source code for each R version. # We just need the files PACKAGES and PACKAGES.gz in CRAN/src/contrib. So I comment out the following line. # rsync -rtlzv --delete cran.r-project.org::CRAN/src/ ~/Rmirror/CRAN/src/ mkdir -p ~/Rmirror/CRAN/src/contrib rsync -rtlzv --delete cran.r-project.org::CRAN/src/contrib/PACKAGES ~/Rmirror/CRAN/src/contrib/ rsync -rtlzv --delete cran.r-project.org::CRAN/src/contrib/PACKAGES.gz ~/Rmirror/CRAN/src/contrib/
And optionally
library(tools)
write_PACKAGES("~/Rmirror/CRAN/bin/windows/contrib/2.15", type="win.binary")
and if we want to get src directory
rsync -rtlzv --delete cran.r-project.org::CRAN/src/contrib/*.tar.gz ~/Rmirror/CRAN/src/contrib/ rsync -rtlzv --delete cran.r-project.org::CRAN/src/contrib/2.15.3 ~/Rmirror/CRAN/src/contrib/
We can use du -h to check the folder size.
For example (as of 1/7/2013),
$ du -k ~/Rmirror --max-depth=1 --exclude ".*" | sort -nr | cut -f2 | xargs -d '\n' du -sh 30G /home/brb/Rmirror 28G /home/brb/Rmirror/Bioc 2.7G /home/brb/Rmirror/CRAN
To create Bioconductor repository
Dry run
rsync -avn bioconductor.org::2.11 > biocdryrun.txt
Then creates directories before running rsync.
cd mkdir -p ~/Rmirror/Bioc wget -N http://www.bioconductor.org/biocLite.R -P ~/Rmirror/Bioc
where -N is to overwrite original file if the size or timestamp change and -P in wget means an output directory, not a file name.
Optionally, we can add the following in order to see the Bioconductor front page.
rsync -zrtlv --delete bioconductor.org::2.11/BiocViews.html ~/Rmirror/Bioc/packages/2.11/ rsync -zrtlv --delete bioconductor.org::2.11/index.html ~/Rmirror/Bioc/packages/2.11/
The software part (aka bioc directory) installation:
cd mkdir -p ~/Rmirror/Bioc/packages/2.11/bioc/bin/windows mkdir -p ~/Rmirror/Bioc/packages/2.11/bioc/src rsync -zrtlv --delete bioconductor.org::2.11/bioc/bin/windows/ ~/Rmirror/Bioc/packages/2.11/bioc/bin/windows # Either rsync whole src directory or just essential files # rsync -zrtlv --delete bioconductor.org::2.11/bioc/src/ ~/Rmirror/Bioc/packages/2.11/bioc/src rsync -zrtlv --delete bioconductor.org::2.11/bioc/src/contrib/PACKAGES ~/Rmirror/Bioc/packages/2.11/bioc/src/contrib/ rsync -zrtlv --delete bioconductor.org::2.11/bioc/src/contrib/PACKAGES.gz ~/Rmirror/Bioc/packages/2.11/bioc/src/contrib/ # Optionally the html part mkdir -p ~/Rmirror/Bioc/packages/2.11/bioc/html rsync -zrtlv --delete bioconductor.org::2.11/bioc/html/ ~/Rmirror/Bioc/packages/2.11/bioc/html mkdir -p ~/Rmirror/Bioc/packages/2.11/bioc/vignettes rsync -zrtlv --delete bioconductor.org::2.11/bioc/vignettes/ ~/Rmirror/Bioc/packages/2.11/bioc/vignettes mkdir -p ~/Rmirror/Bioc/packages/2.11/bioc/news rsync -zrtlv --delete bioconductor.org::2.11/bioc/news/ ~/Rmirror/Bioc/packages/2.11/bioc/news mkdir -p ~/Rmirror/Bioc/packages/2.11/bioc/licenses rsync -zrtlv --delete bioconductor.org::2.11/bioc/licenses/ ~/Rmirror/Bioc/packages/2.11/bioc/licenses mkdir -p ~/Rmirror/Bioc/packages/2.11/bioc/manuals rsync -zrtlv --delete bioconductor.org::2.11/bioc/manuals/ ~/Rmirror/Bioc/packages/2.11/bioc/manuals mkdir -p ~/Rmirror/Bioc/packages/2.11/bioc/readmes rsync -zrtlv --delete bioconductor.org::2.11/bioc/readmes/ ~/Rmirror/Bioc/packages/2.11/bioc/readmes
and annotation (aka data directory) part:
mkdir -p ~/Rmirror/Bioc/packages/2.11/data/annotation/bin/windows mkdir -p ~/Rmirror/Bioc/packages/2.11/data/annotation/src/contrib # one line for each of the following rsync -zrtlv --delete bioconductor.org::2.11/data/annotation/bin/windows/ ~/Rmirror/Bioc/packages/2.11/data/annotation/bin/windows rsync -zrtlv --delete bioconductor.org::2.11/data/annotation/src/contrib/PACKAGES ~/Rmirror/Bioc/packages/2.11/data/annotation/src/contrib/ rsync -zrtlv --delete bioconductor.org::2.11/data/annotation/src/contrib/PACKAGES.gz ~/Rmirror/Bioc/packages/2.11/data/annotation/src/contrib/
and experiment directory:
mkdir -p ~/Rmirror/Bioc/packages/2.11/data/experiment/bin/windows/contrib/2.15 mkdir -p ~/Rmirror/Bioc/packages/2.11/data/experiment/src/contrib # one line for each of the following # Note that we are cheating by only downloading PACKAGES and PACKAGES.gz files rsync -zrtlv --delete bioconductor.org::2.11/data/experiment/bin/windows/contrib/2.15/PACKAGES ~/Rmirror/Bioc/packages/2.11/data/experiment/bin/windows/contrib/2.15/ rsync -zrtlv --delete bioconductor.org::2.11/data/experiment/bin/windows/contrib/2.15/PACKAGES.gz ~/Rmirror/Bioc/packages/2.11/data/experiment/bin/windows/contrib/2.15/ rsync -zrtlv --delete bioconductor.org::2.11/data/experiment/src/contrib/PACKAGES ~/Rmirror/Bioc/packages/2.11/data/experiment/src/contrib/ rsync -zrtlv --delete bioconductor.org::2.11/data/experiment/src/contrib/PACKAGES.gz ~/Rmirror/Bioc/packages/2.11/data/experiment/src/contrib/
and extra directory:
mkdir -p ~/Rmirror/Bioc/packages/2.11/extra/bin/windows/contrib/2.15 mkdir -p ~/Rmirror/Bioc/packages/2.11/extra/src/contrib # one line for each of the following # Note that we are cheating by only downloading PACKAGES and PACKAGES.gz files rsync -zrtlv --delete bioconductor.org::2.11/extra/bin/windows/contrib/2.15/PACKAGES ~/Rmirror/Bioc/packages/2.11/extra/bin/windows/contrib/2.15/ rsync -zrtlv --delete bioconductor.org::2.11/extra/bin/windows/contrib/2.15/PACKAGES.gz ~/Rmirror/Bioc/packages/2.11/extra/bin/windows/contrib/2.15/ rsync -zrtlv --delete bioconductor.org::2.11/extra/src/contrib/PACKAGES ~/Rmirror/Bioc/packages/2.11/extra/src/contrib/ rsync -zrtlv --delete bioconductor.org::2.11/extra/src/contrib/PACKAGES.gz ~/Rmirror/Bioc/packages/2.11/extra/src/contrib/
sync Bioconductor software packages
To keep a copy of the bioc/source (software packages) code only,
$ mkdir -p ~/bioc_release/bioc/ $ rsync -zrtlv --delete master.bioconductor.org::release/bioc/src ~/bioc_release/bioc/ $ du -h ~/bioc_release/bioc/ # 20GB, 1565 items, Bioc 3.7
Note -z - compress file data during the transfer, -t - preserve modification times, -l copy symbolic links as symbolic links. The option -zrtlv can be replaced by the common options -avz.
To get the old versions of a packages (after the release of a version of Bioconductor), check out the Archive folder.
Now we can create a cron job to do sync. Note my observation is Bioconductor has a daily update around 10:45AM. So I set time at 11:00AM.
echo "00 11 * * * rsync -avz --delete master.bioconductor.org::release/bioc/src ~/bioc_release/bioc/" >> \ ~/Documents/cronjob # everyday at 6am & 1pm crontab ~/Documents/cronjob crontab -l
To test local repository
Create soft links in Apache server
su ln -s /home/brb/Rmirror/CRAN /var/www/html/CRAN ln -s /home/brb/Rmirror/Bioc /var/www/html/Bioc ls -l /var/www/html
The soft link mode should be 777.
To test CRAN
Replace the host name arraytools.no-ip.org by IP address 10.133.2.111 if necessary.
r <- getOption("repos"); r["CRAN"] <- "http://arraytools.no-ip.org/CRAN"
options(repos=r)
install.packages("glmnet")
We can test if the backup server is working or not by installing a package which was removed from the CRAN. For example, 'ForImp' was removed from CRAN in 11/8/2012, but I still a local copy built on R 2.15.2 (run rsync on 11/6/2012).
r <- getOption("repos"); r["CRAN"] <- "http://cran.r-project.org"
r <- c(r, BRB='http://arraytools.no-ip.org/CRAN')
# CRAN CRANextra BRB
# "http://cran.r-project.org" "http://www.stats.ox.ac.uk/pub/RWin" "http://arraytools.no-ip.org/CRAN"
options(repos=r)
install.packages('ForImp')
Note by default, CRAN mirror is selected interactively.
> getOption("repos")
CRAN CRANextra
"@CRAN@" "http://www.stats.ox.ac.uk/pub/RWin"
To test Bioconductor
# CRAN part:
r <- getOption("repos"); r["CRAN"] <- "http://arraytools.no-ip.org/CRAN"
options(repos=r)
# Bioconductor part:
options("BioC_mirror" = "http://arraytools.no-ip.org/Bioc")
source("http://bioconductor.org/biocLite.R")
# This source biocLite.R line can be placed either before or after the previous 2 lines
biocLite("aCGH")
If there is a connection problem, check folder attributes.
chmod -R 755 ~/CRAN/bin
- Note that if a binary package was created for R 2.15.1, then it can be installed under R 2.15.1 but not R 2.15.2. The R console will show package xxx is not available (for R version 2.15.2).
- For binary installs, the function also checks for the availability of a source package on the same repository, and reports if the source package has a later version, or is available but no binary version is.
So for example, if the mirror does not have contents under src directory, we need to run the following line in order to successfully run install.packages() function.
options(install.packages.check.source = "no")
- If we only mirror the essential directories, we can run biocLite() successfully. However, the R console will give some warning
> biocLite("aCGH")
BioC_mirror: http://arraytools.no-ip.org/Bioc
Using Bioconductor version 2.11 (BiocInstaller 1.8.3), R version 2.15.
Installing package(s) 'aCGH'
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/data/experiment/src/contrib
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/extra/src/contrib
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/data/experiment/bin/windows/contrib/2.15
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/extra/bin/windows/contrib/2.15
trying URL 'http://arraytools.no-ip.org/Bioc/packages/2.11/bioc/bin/windows/contrib/2.15/aCGH_1.36.0.zip'
Content type 'application/zip' length 2431158 bytes (2.3 Mb)
opened URL
downloaded 2.3 Mb
package ‘aCGH’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\limingc\AppData\Local\Temp\Rtmp8IGGyG\downloaded_packages
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/data/experiment/bin/windows/contrib/2.15
Warning: unable to access index for repository http://arraytools.no-ip.org/Bioc/packages/2.11/extra/bin/windows/contrib/2.15
> library()
CRAN repository directory structure
The information below is specific to R 2.15.2. There are linux and macosx subdirecotries whenever there are windows subdirectory.
bin/winows/contrib/2.15 src/contrib /contrib/2.15.2 /contrib/Archive web/checks /dcmeta /packages /views
A clickable map [1]
CRAN package download statistics from RStudio
- Daily download statistics http://cran-logs.rstudio.com/. Note the page is split into 'package' download and 'R' download. It tracks
- Package: date, time, size, r_version, r_arch, r_os, package, version, country, ip_id.
- R: date, time, size, R version, os (win/src/osx), county, ip_id (reset daily).
- https://www.r-bloggers.com/finally-tracking-cran-packages-downloads/. The code still works.
- https://strengejacke.wordpress.com/2015/03/07/cran-download-statistics-of-any-packages-rstats/
Bioconductor package download statistics
http://bioconductor.org/packages/stats/
Bioconductor repository directory structure
The information below is specific to Bioc 2.11 (R 2.15). There are linux and macosx subdirecotries whenever there are windows subdirectory.
bioc/bin/windows/contrib/2.15
/html
/install
/license
/manuals
/news
/src
/vignettes
data/annotation/bin/windows/contrib/2.15
/html
/licenses
/manuals
/src
/vignettes
/experiment/bin/windows/contrib/2.15
/html
/manuals
/src/contrib
/vignettes
extra/bin/windows/contrib
/html
/src
/vignettes
List all R packages from CRAN/Bioconductor
Check my daily result based on R 2.15 and Bioc 2.11 in [2]
See METACRAN for packages hosted on CRAN. The 'https://github.com/metacran/PACKAGES' file contains the latest update.
r-hub: the everything-builder the R community needs
https://github.com/r-hub/proposal
Introducing R-hub, the R package builder service
- https://www.rstudio.com/resources/videos/r-hub-overview/
- http://blog.revolutionanalytics.com/2016/10/r-hub-public-beta.html
Parallel Computing
- Example code for the book Parallel R by McCallum and Weston.
- A gentle introduction to parallel computing in R
- An introduction to distributed memory parallelism in R and C
- Processing: When does it worth?
Security warning from Windows/Mac
It seems it is safe to choose 'Cancel' when Windows Firewall tried to block R program when we use makeCluster() to create a socket cluster.
library(parallel)
cl <- makeCluster(2)
clusterApply(cl, 1:2, get("+"), 3)
stopCluster(cl)
If we like to see current firewall settings, just click Windows Start button, search 'Firewall' and choose 'Windows Firewall with Advanced Security'. In the 'Inbound Rules', we can see what programs (like, R for Windows GUI front-end, or Rserve) are among the rules. These rules are called 'private' in the 'Profile' column. Note that each of them may appear twice because one is 'TCP' protocol and the other one has a 'UDP' protocol.
Detect number of cores
parallel::detectCores()
Don't use the default option getOption("mc.cores", 2L) (PS it only returns 2.) in mclapply() unless you are a developer for a package.
However, it is a different story when we run the R code in HPC cluster. Read the discussion Whether to use the detectCores function in R to specify the number of cores for parallel processing?
On NIH's biowulf, even I specify an interactive session with 4 cores, the parallel::detectCores() function returns 56. This number is the same as the output from the bash command grep processor /proc/cpuinfo or (better) lscpu. The free -hm also returns a full 125GB size instead of my requested size (4GB by default).
parallel package
Parallel package was included in R 2.14.0. It is derived from the snow and multicore packages and provides many of the same functions as those packages.
The parallel package provides several *apply functions for R users to quickly modify their code using parallel computing.
- makeCluster(makePSOCKcluster, makeForkCluster), stopCluster. Other cluster types are passed to package snow.
- clusterCall, clusterEvalQ: source R files and/or load libraries
- clusterSplit
- clusterApply, clusterApplyLB (vs the foreach package)
- clusterExport: export variables
- clusterMap
- parLapply, parSapply, parApply, parRapply, parCapply. Note that
- parSapply() can be used to as a parallel version of the replicate() function. See this example.
- An iteration parameter needs to be added to the first parameter of the main function.
- parLapplyLB, parSapplyLB (load balance version)
- clusterSetRNGStream, nextRNGStream, nextRNGSubStream
Examples (See ?clusterApply)
library(parallel)
cl <- makeCluster(2, type = "SOCK")
clusterApply(cl, 1:2, function(x) x*3) # OR clusterApply(cl, 1:2, get("*"), 3)
# [[1]]
# [1] 3
#
# [[2]]
# [1] 6
parSapply(cl, 1:20, get("+"), 3)
# [1] 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23
stopCluster(cl)
An example of using clusterCall() or clusterEvalQ()
library(parallel)
cl <- makeCluster(4)
clusterCall(cl, function() {
source("test.R")
})
# clusterEvalQ(cl, {
# source("test.R")
# })
## do some parallel work
stopCluster(cl)
snow package
Supported cluster types are "SOCK", "PVM", "MPI", and "NWS".
multicore package
This package is removed from CRAN.
Consider using package ‘parallel’ instead.
foreach package
This package depends on one of the following
- doParallel - Foreach parallel adaptor for the parallel package
- doSNOW - Foreach parallel adaptor for the snow package
- doMC - Foreach parallel adaptor for the multicore package. Used in glmnet vignette.
- doMPI - Foreach parallel adaptor for the Rmpi package
- doRedis - Foreach parallel adapter for the rredis package
as a backend.
library(foreach) library(doParallel) m <- matrix(rnorm(9), 3, 3) cl <- makeCluster(2, type = "SOCK") registerDoParallel(cl) # register the parallel backend with the foreach package foreach(i=1:nrow(m), .combine=rbind) %dopar% (m[i,] / mean(m[i,])) stopCluster(cl)
See also this post Updates to the foreach package and its friends on Oct 2015.
combine list of lists
- .combine argument https://stackoverflow.com/questions/27279164/output-list-of-two-rbinded-data-frames-with-foreach-in-r
- Merge lists by mapply() or base::Map()
comb <- function(...) {
mapply('cbind', ..., SIMPLIFY=FALSE)
}
library(foreach)
library(doParallel)
cl <- makeCluster(2)
registerDoParallel(cl) # register the parallel backend with the foreach package
m <- rbind(rep(1,3), rep(2,3))
# nrow(m) can represents number of permutations (2 in this toy example)
tmp <- foreach(i=1:nrow(m)) %dopar% {
a <- m[i, ]
b <- a * 10
list(a, b)
}; tmp
# [[1]]
# [[1]][[1]]
# [1] 1 1 1
#
# [[1]][[2]]
# [1] 10 10 10
#
#
# [[2]]
# [[2]][[1]]
# [1] 2 2 2
#
# [[2]][[2]]
# [1] 20 20 20
foreach(i=1:nrow(m), .combine = "comb") %dopar% {
a <- m[i,]
b <- a * 10
list(a, b)
}
# [[1]]
# [,1] [,2]
# [1,] 1 2
# [2,] 1 2
# [3,] 1 2
#
# [[2]]
# [,1] [,2]
# [1,] 10 20
# [2,] 10 20
# [3,] 10 20
stopCluster(cl)
Replacing double loops
- https://stackoverflow.com/questions/30927693/how-can-i-parallelize-a-double-for-loop-in-r
- http://www.exegetic.biz/blog/2013/08/the-wonders-of-foreach/
library(foreach)
library(doParallel)
nc <- 4
nr <- 2
cores=detectCores()
cl <- makeCluster(cores[1]-1)
registerDoParallel(cl)
# set.seed(1234) # not work
# set.seed(1234, "L'Ecuyer-CMRG") # not work either
# library("doRNG")
# registerDoRNG(seed = 1985) # not work with nested foreach
# Error in list(e1 = list(args = (1:nr)(), argnames = "i", evalenv = <environment>, :
# nested/conditional foreach loops are not supported yet.
m <- foreach (i = 1:nr, .combine='rbind') %:% # nesting operator
foreach (j = 1:nc) %dopar% {
rnorm(1, i*5, j) # code to parallelise
}
m
stopCluster(cl)
Note that since the random seed (see the next session) does not work on nested loop, it is better to convert nested loop (two indices) to a single loop (one index).
set seed and doRNG package
- Vignette, Documentation
- doRNG package example
- How to set seed for random simulations with foreach and doMC packages?
- Use clusterSetRNGStream() from the parallel package; see How-to go parallel in R – basics + tips
- http://www.stat.colostate.edu/~scharfh/CSP_parallel/handouts/foreach_handout.html#random-numbers
library("doRNG") # doRNG does not need to be loaded after doParallel library("doParallel") cl <- makeCluster(2) registerDoParallel(cl) registerDoRNG(seed = 1234) # works for a single loop m1 <- foreach(i = 1:5, .combine = 'c') %dopar% rnorm(1) registerDoRNG(seed = 1234) m2 <- foreach(i = 1:5, .combine = 'c') %dopar% rnorm(1) identical(m1, m2) stopCluster(cl) attr(m1, "rng") <- NULL # remove rng attribute - Another way to use the seed is to supply .options.RNG in foreach() function.
r1 <- foreach(i=1:4, .options.RNG=1234) %dorng% { runif(1) }
Export libraries and variables
clusterEvalQ(cl, {
library(biospear)
library(glmnet)
library(survival)
})
clusterExport(cl, list("var1", "foo2"))
Summary the result
foreach returns the result in a list. For example, if each component is a matrix we can use
- Reduce("+", res)/length(res) # Reduce("+", res, na.rm = TRUE) not working
- apply(simplify2array(res), 1:2, mean, na.rm = TRUE)
to get the average of matrices over the list.
Read a list files
Reading List Faster With parallel, doParallel, and pbapply
snowfall package
Rmpi package
Some examples/tutorials
- http://trac.nchc.org.tw/grid/wiki/R-MPI_Install
- http://www.arc.vt.edu/resources/software/r/index.php
- https://www.sharcnet.ca/help/index.php/Using_R_and_MPI
- http://math.acadiau.ca/ACMMaC/Rmpi/examples.html
- http://www.umbc.edu/hpcf/resources-tara/how-to-run-R.html
- Ryan Rosario
- http://pj.freefaculty.org/guides/Rcourse/parallel-1/parallel-1.pdf
- * http://biowulf.nih.gov/apps/R.html
OpenMP
- R and openMP: boosting compiled code on multi-core cpu-s from parallelr.com.
BiocParallel
RcppParallel
future & future.apply package
- Asynchronous and Distributed Programming in R with the Future Package
- Parallelize Any Base R Apply Function
Apache Spark
Microsoft R Server
- Microsoft R Server (not Microsoft R Open)
GPU
- GPU Programming for All with ‘gpuR from parallelr.com. The gpuR is available on CRAN.
- gputools
Threads
- Rdsm package
- (A Very) Experimental Threading in R and a post from Mad Scientist
Benchmark
Are parallel simulations in the cloud worth it? Benchmarking my MBP vs my Workstation vs Amazon EC2
R functions to run timing
# Method 1 system.time( invisible(rnorm(10000))) # Method 2 btime <- Sys.time() invisible(rnorm(10000)) Sys.time() - btime
Cloud Computing
Install R on Amazon EC2
http://randyzwitch.com/r-amazon-ec2/
Bioconductor on Amazon EC2
http://www.bioconductor.org/help/bioconductor-cloud-ami/
Big Data Analysis
Useful R packages
RInside
- http://dirk.eddelbuettel.com/code/rinside.html
- http://dirk.eddelbuettel.com/papers/rfinance2010_rcpp_rinside_tutorial_handout.pdf
Ubuntu
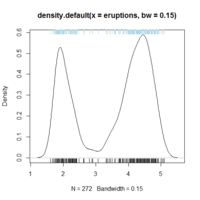
With RInside, R can be embedded in a graphical application. For example, $HOME/R/x86_64-pc-linux-gnu-library/3.0/RInside/examples/qt directory includes source code of a Qt application to show a kernel density plot with various options like kernel functions, bandwidth and an R command text box to generate the random data. See my demo on Youtube. I have tested this qtdensity example successfully using Qt 4.8.5.
- Follow the instruction cairoDevice to install required libraries for cairoDevice package and then cairoDevice itself.
- Install Qt. Check 'qmake' command becomes available by typing 'whereis qmake' or 'which qmake' in terminal.
- Open Qt Creator from Ubuntu start menu/Launcher. Open the project file $HOME/R/x86_64-pc-linux-gnu-library/3.0/RInside/examples/qt/qtdensity.pro in Qt Creator.
- Under Qt Creator, hit 'Ctrl + R' or the big green triangle button on the lower-left corner to build/run the project. If everything works well, you shall see the interactive program qtdensity appears on your desktop.
With RInside + Wt web toolkit installed, we can also create a web application. To demonstrate the example in examples/wt directory, we can do
cd ~/R/x86_64-pc-linux-gnu-library/3.0/RInside/examples/wt make sudo ./wtdensity --docroot . --http-address localhost --http-port 8080
Then we can go to the browser's address bar and type http://localhost:8080 to see how it works (a screenshot is in here).
Windows 7
To make RInside works on Windows OS, try the following
- Make sure R is installed under C:\ instead of C:\Program Files if we don't want to get an error like g++.exe: error: Files/R/R-3.0.1/library/RInside/include: No such file or directory.
- Install RTools
- Instal RInside package from source (the binary version will give an error )
- Create a DOS batch file containing necessary paths in PATH environment variable
@echo off set PATH=C:\Rtools\bin;c:\Rtools\gcc-4.6.3\bin;%PATH% set PATH=C:\R\R-3.0.1\bin\i386;%PATH% set PKG_LIBS=`Rscript -e "Rcpp:::LdFlags()"` set PKG_CPPFLAGS=`Rscript -e "Rcpp:::CxxFlags()"` set R_HOME=C:\R\R-3.0.1 echo Setting environment for using R cmd
In the Windows command prompt, run
cd C:\R\R-3.0.1\library\RInside\examples\standard make -f Makefile.win
Now we can test by running any of executable files that make generates. For example, rinside_sample0.
rinside_sample0
As for the Qt application qdensity program, we need to make sure the same version of MinGW was used in building RInside/Rcpp and Qt. See some discussions in
- http://stackoverflow.com/questions/12280707/using-rinside-with-qt-in-windows
- http://www.mail-archive.com/[email protected]/msg04377.html
So the Qt and Wt web tool applications on Windows may or may not be possible.
GUI
Qt and R
- http://cran.r-project.org/web/packages/qtbase/index.html QtDesigner is such a tool, and its output is compatible with the qtbase R package
- http://qtinterfaces.r-forge.r-project.org
tkrplot
On Ubuntu, we need to install tk packages, such as by
sudo apt-get install tk-dev
reticulate - Interface to 'Python'
- https://cran.r-project.org/web/packages/reticulate/index.html
- R or Python? Why not both? Using Anaconda Python within R with {reticulate}
Hadoop (eg ~100 terabytes)
See also HighPerformanceComputing
- RHadoop
- Hive
- MapReduce. Introduction by Linux Journal.
- http://www.techspritz.com/category/tutorials/hadoopmapredcue/ Single node or multinode cluster setup using Ubuntu with VirtualBox (Excellent)
- Running Hadoop on Ubuntu Linux (Single-Node Cluster)
- Ubuntu 12.04 http://www.youtube.com/watch?v=WN2tJk_oL6E and instruction
- Linux Mint http://blog.hackedexistence.com/installing-hadoop-single-node-on-linux-mint
- http://www.r-bloggers.com/search/hadoop
RHadoop
- RDataMining.com based on Mac.
- Ubuntu 12.04 - Crishantha.com, nikhilshah123sh.blogspot.com.Bighadoop.wordpress contains an example.
- RapReduce in R by RevolutionAnalytics with a few examples.
- https://twitter.com/hashtag/rhadoop
- Bigd8ta.com based on Ubuntu 14.04.
Snowdoop: an alternative to MapReduce algorithm
- http://matloff.wordpress.com/2014/11/26/how-about-a-snowdoop-package/
- http://matloff.wordpress.com/2014/12/26/snowdooppartools-update/comment-page-1/#comment-665
XML
On Ubuntu, we need to install libxml2-dev before we can install XML package.
sudo apt-get update sudo apt-get install libxml2-dev
On CentOS,
yum -y install libxml2 libxml2-devel
XML
- http://giventhedata.blogspot.com/2012/06/r-and-web-for-beginners-part-ii-xml-in.html. It gave an example of extracting the XML-values from each XML-tag for all nodes and save them in a data frame using xmlSApply().
- http://www.quantumforest.com/2011/10/reading-html-pages-in-r-for-text-processing/
- https://tonybreyal.wordpress.com/2011/11/18/htmltotext-extracting-text-from-html-via-xpath/
- https://www.tutorialspoint.com/r/r_xml_files.htm
- https://www.datacamp.com/community/tutorials/r-data-import-tutorial#xml
- Extracting data from XML PubMed and Zillow are used to illustrate. xmlTreeParse(), xmlRoot(), xmlName() and xmlSApply().
- https://yihui.name/en/2010/10/grabbing-tables-in-webpages-using-the-xml-package/
library(XML)
# Read and parse HTML file
doc.html = htmlTreeParse('http://apiolaza.net/babel.html', useInternal = TRUE)
# Extract all the paragraphs (HTML tag is p, starting at
# the root of the document). Unlist flattens the list to
# create a character vector.
doc.text = unlist(xpathApply(doc.html, '//p', xmlValue))
# Replace all by spaces
doc.text = gsub('\n', ' ', doc.text)
# Join all the elements of the character vector into a single
# character string, separated by spaces
doc.text = paste(doc.text, collapse = ' ')
This post http://stackoverflow.com/questions/25315381/using-xpathsapply-to-scrape-xml-attributes-in-r can be used to monitor new releases from github.com.
> library(RCurl) # getURL()
> library(XML) # htmlParse and xpathSApply
> xData <- getURL("https://github.com/alexdobin/STAR/releases")
> doc = htmlParse(xData)
> plain.text <- xpathSApply(doc, "//span[@class='css-truncate-target']", xmlValue)
# I look at the source code and search 2.5.3a and find the tag as
# <span class="css-truncate-target">2.5.3a</span>
> plain.text
[1] "2.5.3a" "2.5.2b" "2.5.2a" "2.5.1b" "2.5.1a"
[6] "2.5.0c" "2.5.0b" "STAR_2.5.0a" "STAR_2.4.2a" "STAR_2.4.1d"
>
> # try bwa
> > xData <- getURL("https://github.com/lh3/bwa/releases")
> doc = htmlParse(xData)
> xpathSApply(doc, "//span[@class='css-truncate-target']", xmlValue)
[1] "v0.7.15" "v0.7.13"
> # try picard
> xData <- getURL("https://github.com/broadinstitute/picard/releases")
> doc = htmlParse(xData)
> xpathSApply(doc, "//span[@class='css-truncate-target']", xmlValue)
[1] "2.9.1" "2.9.0" "2.8.3" "2.8.2" "2.8.1" "2.8.0" "2.7.2" "2.7.1" "2.7.0"
[10] "2.6.0"
This method can be used to monitor new tags/releases from some projects like Cura, BWA, Picard, STAR. But for some projects like sratools the class attribute in the span element ("css-truncate-target") can be different (such as "tag-name").
xmlview
RCurl
On Ubuntu, we need to install the packages (the first one is for XML package that RCurl suggests)
# Test on Ubuntu 14.04 sudo apt-get install libxml2-dev sudo apt-get install libcurl4-openssl-dev
Scrape google scholar results
No google ID is required
Seems not work
Error in data.frame(footer = xpathLVApply(doc, xpath.base, "/font/span[@class='gs_fl']", : arguments imply differing number of rows: 2, 0
devtools
devtools package depends on Curl.
# Test on Ubuntu 14.04 sudo apt-get install libcurl4-openssl-dev
httr
httr imports curl, jsonlite, mime, openssl and R6 packages.
When I tried to install httr package, I got an error and some message:
Configuration failed because openssl was not found. Try installing: * deb: libssl-dev (Debian, Ubuntu, etc) * rpm: openssl-devel (Fedora, CentOS, RHEL) * csw: libssl_dev (Solaris) * brew: openssl (Mac OSX) If openssl is already installed, check that 'pkg-config' is in your PATH and PKG_CONFIG_PATH contains a openssl.pc file. If pkg-config is unavailable you can set INCLUDE_DIR and LIB_DIR manually via: R CMD INSTALL --configure-vars='INCLUDE_DIR=... LIB_DIR=...' -------------------------------------------------------------------- ERROR: configuration failed for package ‘openssl’
It turns out after I run sudo apt-get install libssl-dev in the terminal (Debian), it would go smoothly with installing httr package. Nice httr!
Real example: see this post. Unfortunately I did not get a table result; I only get an html file (R 3.2.5, httr 1.1.0 on Ubuntu and Debian).
Since httr package was used in many other packages, take a look at how others use it. For example, aRxiv package.
curl
curl is independent of RCurl package.
- http://cran.r-project.org/web/packages/curl/vignettes/intro.html
- https://www.opencpu.org/posts/curl-release-0-8/
library(curl)
h <- new_handle()
handle_setform(h,
name="aaa", email="bbb"
)
req <- curl_fetch_memory("http://localhost/d/phpmyql3_scripts/ch02/form2.html", handle = h)
rawToChar(req$content)
rOpenSci packages
rOpenSci contains packages that allow access to data repositories through the R statistical programming environment
remotes
DirichletMultinomial
On Ubuntu, we do
sudo apt-get install libgsl0-dev
Create GUI
gWidgets
GenOrd: Generate ordinal and discrete variables with given correlation matrix and marginal distributions
rjson
http://heuristically.wordpress.com/2013/05/20/geolocate-ip-addresses-in-r/
RJSONIO
Accessing Bitcoin Data with R
http://blog.revolutionanalytics.com/2015/11/accessing-bitcoin-data-with-r.html
Plot IP on google map
- http://thebiobucket.blogspot.com/2011/12/some-fun-with-googlevis-plotting-blog.html#more (RCurl, RJONIO, plyr, googleVis)
- http://devblog.icans-gmbh.com/using-the-maxmind-geoip-api-with-r/ (RCurl, RJONIO, maps)
- http://cran.r-project.org/web/packages/geoPlot/index.html (geoPlot package (deprecated as 8/12/2013))
- http://archive09.linux.com/feature/135384 (Not R) ApacheMap
- http://batchgeo.com/features/geolocation-ip-lookup/ (Not R) (Enter a spreadsheet of adress, city, zip or a column of IPs and it will show the location on google map)
- http://code.google.com/p/apachegeomap/
The following example is modified from the first of above list.
require(RJSONIO) # fromJSON
require(RCurl) # getURL
temp = getURL("https://gist.github.com/arraytools/6743826/raw/23c8b0bc4b8f0d1bfe1c2fad985ca2e091aeb916/ip.txt",
ssl.verifypeer = FALSE)
ip <- read.table(textConnection(temp), as.is=TRUE)
names(ip) <- "IP"
nr = nrow(ip)
Lon <- as.numeric(rep(NA, nr))
Lat <- Lon
Coords <- data.frame(Lon, Lat)
ip2coordinates <- function(ip) {
api <- "http://freegeoip.net/json/"
get.ips <- getURL(paste(api, URLencode(ip), sep=""))
# result <- ldply(fromJSON(get.ips), data.frame)
result <- data.frame(fromJSON(get.ips))
names(result)[1] <- "ip.address"
return(result)
}
for (i in 1:nr){
cat(i, "\n")
try(
Coords[i, 1:2] <- ip2coordinates(ip$IP[i])[c("longitude", "latitude")]
)
}
# append to log-file:
logfile <- data.frame(ip, Lat = Coords$Lat, Long = Coords$Lon,
LatLong = paste(round(Coords$Lat, 1), round(Coords$Lon, 1), sep = ":"))
log_gmap <- logfile[!is.na(logfile$Lat), ]
require(googleVis) # gvisMap
gmap <- gvisMap(log_gmap, "LatLong",
options = list(showTip = TRUE, enableScrollWheel = TRUE,
mapType = 'hybrid', useMapTypeControl = TRUE,
width = 1024, height = 800))
plot(gmap)
The plot.gvis() method in googleVis packages also teaches the startDynamicHelp() function in the tools package, which was used to launch a http server. See Jeffrey Horner's note about deploying Rook App.
Map
leaflet
- rstudio.github.io/leaflet/#installation-and-use
- https://metvurst.wordpress.com/2015/07/24/mapview-basic-interactive-viewing-of-spatial-data-in-r-6/
choroplethr
- http://blog.revolutionanalytics.com/2014/01/easy-data-maps-with-r-the-choroplethr-package-.html
- http://www.arilamstein.com/blog/2015/06/25/learn-to-map-census-data-in-r/
- http://www.arilamstein.com/blog/2015/09/10/user-question-how-to-add-a-state-border-to-a-zip-code-map/
ggplot2
How to make maps with Census data in R
googleVis
See an example from RJSONIO above.
googleAuthR
Create R functions that interact with OAuth2 Google APIs easily, with auto-refresh and Shiny compatibility.
gtrendsR - Google Trends
- Download and plot Google Trends data with R
- Analyzing Google Trends Data in R
- microarray analysis from 2004-04-01
- ngs next generation sequencing from 2004-04-01
- dna sequencing from 2004-01-01.
- rna sequencing from 2004-01-01. It can be seen RNA sequencing >> DNA sequencing.
- Python vs R – Who Is Really Ahead in Data Science, Machine Learning? and The Incredible Growth of Python by David Robinson
quantmod
Maintaining a database of price files in R. It consists of 3 steps.
- Initial data downloading
- Update existing data
- Create a batch file
Rcpp
- Discussion archive
- (Video) Extending R with C++: A Brief Introduction to Rcpp
- C++14, R and Travis -- A useful hack
It may be necessary to install dependency packages for RcppEigen.
sudo apt-get install libblas-dev liblapack-dev sudo apt-get install gfortran
Speed Comparison
- A comparison of high-performance computing techniques in R. It compares Rcpp to an R looping operator (like mapply), a parallelized version of a looping operator (like mcmapply), explicit parallelization, via the parallel package or the ParallelR suite.
- In the following example, C++ avoids the overhead of creating an intermediate object (eg vector of the same length as the original vector). The c++ uses an intermediate scalar. So C++ wins R over memory management in this case.
# http://blog.mckuhn.de/2016/03/avoiding-unnecessary-memory-allocations.html library(Rcpp) `%count<%` <- cppFunction(' size_t count_less(NumericVector x, NumericVector y) { const size_t nx = x.size(); const size_t ny = y.size(); if (nx > 1 & ny > 1) stop("Only one parameter can be a vector!"); size_t count = 0; if (nx == 1) { double c = x[0]; for (int i = 0; i < ny; i++) count += c < y[i]; } else { double c = y[0]; for (int i = 0; i < nx; i++) count += x[i] < c; } return count; } ') set.seed(42) N <- 10^7 v <- runif(N, 0, 10000) # Testing on my ODroid xu4 running ubuntu 15.10 system.time(sum(v < 5000)) # user system elapsed # 1.135 0.305 1.453 system.time(v %count<% 5000) # user system elapsed # 0.535 0.000 0.540 - Why R for data science – and not Python?
library(Rcpp) bmi_R <- function(weight, height) { weight / (height * height) } bmi_R(80, 1.85) # body mass index of person with 80 kg and 185 cm ## [1] 23.37473 cppFunction(" float bmi_cpp(float weight, float height) { return weight / (height * height); } ") bmi_cpp(80, 1.85) # same with cpp function ## [1] 23.37473 - Boost the speed of R calls from Rcpp
Use Rcpp in RStudio
RStudio makes it easy to use Rcpp package.
Open RStudio, click New File -> C++ File. It will create a C++ template on the RStudio editor
#include <Rcpp.h>
using namespace Rcpp;
// Below is a simple example of exporting a C++ function to R. You can
// source this function into an R session using the Rcpp::sourceCpp
// function (or via the Source button on the editor toolbar)
// For more on using Rcpp click the Help button on the editor toolbar
// [[Rcpp::export]]
int timesTwo(int x) {
return x * 2;
}
Now in R console, type
library(Rcpp)
sourceCpp("~/Downloads/timesTwo.cpp")
timesTwo(9)
# [1] 18
See more examples on http://adv-r.had.co.nz/Rcpp.html and Calculating a fuzzy kmeans membership matrix
If we wan to test Boost library, we can try it in RStudio. Consider the following example in stackoverflow.com.
// [[Rcpp::depends(BH)]]
#include <Rcpp.h>
#include <boost/foreach.hpp>
#include <boost/math/special_functions/gamma.hpp>
#define foreach BOOST_FOREACH
using namespace boost::math;
//[[Rcpp::export]]
Rcpp::NumericVector boost_gamma( Rcpp::NumericVector x ) {
foreach( double& elem, x ) {
elem = boost::math::tgamma(elem);
};
return x;
}
Then the R console
boost_gamma(0:10 + 1) # [1] 1 1 2 6 24 120 720 5040 40320 # [10] 362880 3628800 identical( boost_gamma(0:10 + 1), factorial(0:10) ) # [1] TRUE
Example 1. convolution example
First, Rcpp package should be installed (I am working on Linux system). Next we try one example shipped in Rcpp package.
PS. If R was not available in global environment (such as built by ourselves), we need to modify 'Makefile' file by replacing 'R' command with its complete path (4 places).
cd ~/R/x86_64-pc-linux-gnu-library/3.0/Rcpp/examples/ConvolveBenchmarks/ make R
Then type the following in an R session to see how it works. Note that we don't need to issue library(Rcpp) in R.
dyn.load("convolve3_cpp.so")
x <- .Call("convolve3cpp", 1:3, 4:6)
x # 4 13 28 27 18
If we have our own cpp file, we need to use the following way to create dynamic loaded library file. Note that the character (grave accent) ` is not (single quote)'. If you mistakenly use ', it won't work.
export PKG_CXXFLAGS=`Rscript -e "Rcpp:::CxxFlags()"` export PKG_LIBS=`Rscript -e "Rcpp:::LdFlags()"` R CMD SHLIB xxxx.cpp
Example 2. Use together with inline package
library(inline) src <-' Rcpp::NumericVector xa(a); Rcpp::NumericVector xb(b); int n_xa = xa.size(), n_xb = xb.size(); Rcpp::NumericVector xab(n_xa + n_xb - 1); for (int i = 0; i < n_xa; i++) for (int j = 0; j < n_xb; j++) xab[i + j] += xa[i] * xb[j]; return xab; ' fun <- cxxfunction(signature(a = "numeric", b = "numeric"), src, plugin = "Rcpp") fun(1:3, 1:4) # [1] 1 4 10 16 17 12
Example 3. Calling an R function
RcppParallel
caret
- http://topepo.github.io/caret/index.html & https://github.com/topepo/caret/
- https://www.r-project.org/conferences/useR-2013/Tutorials/kuhn/user_caret_2up.pdf
- https://github.com/cran/caret source code mirrored on github
- Cheatsheet https://www.rstudio.com/resources/cheatsheets/
Tool for connecting Excel with R
- https://bert-toolkit.com/
- BERT: a newcomer in the R Excel connection
- http://blog.revolutionanalytics.com/2018/08/how-to-use-r-with-excel.html
Read/Write Excel files package
- http://www.milanor.net/blog/?p=779
- flipAPI. One useful feature of DownloadXLSX, which is not supported by the readxl package, is that it can read Excel files directly from the URL.
- xlsx: depends on Java
- openxlsx: not depend on Java. Depend on zip application. On Windows, it seems to be OK without installing Rtools. But it can not read xls file; it works on xlsx file.
- It can't be used to open .xls or .xlm files.
- When I try the package to read an xlsx file, I got a warning: No data found on worksheet. 6/28/2018
- Use R to write multiple tables to a single Excel file
- readxl: it does not depend on anything although it can only read but not write Excel files. readxl webinar. One advantage of read_excel (as with read_csv in the readr package) is that the data imports into an easy to print object with three attributes a tbl_df, a tbl and a data.frame.
- writexl: zero dependency xlsx writer for R
Tested it on Ubuntu machine with R 3.1.3 using <BRCA.xls> file. Usage:
library(readxl) read_excel(path, sheet = 1, col_names = TRUE, col_types = NULL, na = "", skip = 0)
For the Chromosome column, integer values becomes strings (but converted to double, so 5 becomes 5.000000) or NA (empty on sheets).
> head(read_excel("~/Downloads/BRCA.xls", 4)[ , -9], 3)
UniqueID (Double-click) CloneID UGCluster
1 HK1A1 21652 Hs.445981
2 HK1A2 22012 Hs.119177
3 HK1A4 22293 Hs.501376
Name Symbol EntrezID
1 Catenin (cadherin-associated protein), alpha 1, 102kDa CTNNA1 1495
2 ADP-ribosylation factor 3 ARF3 377
3 Uroporphyrinogen III synthase UROS 7390
Chromosome Cytoband ChimericClusterIDs Filter
1 5.000000 5q31.2 <NA> 1
2 12.000000 12q13 <NA> 1
3 <NA> 10q25.2-q26.3 <NA> 1
The hidden worksheets become visible (Not sure what are those first rows mean in the output).
> excel_sheets("~/Downloads/BRCA.xls")
DEFINEDNAME: 21 00 00 01 0b 00 00 00 02 00 00 00 00 00 00 0d 3b 01 00 00 00 9a 0c 00 00 1a 00
DEFINEDNAME: 21 00 00 01 0b 00 00 00 04 00 00 00 00 00 00 0d 3b 03 00 00 00 9b 0c 00 00 0a 00
DEFINEDNAME: 21 00 00 01 0b 00 00 00 03 00 00 00 00 00 00 0d 3b 02 00 00 00 9a 0c 00 00 06 00
[1] "Experiment descriptors" "Filtered log ratio" "Gene identifiers"
[4] "Gene annotations" "CollateInfo" "GeneSubsets"
[7] "GeneSubsetsTemp"
The Chinese character works too.
> read_excel("~/Downloads/testChinese.xlsx", 1)
中文 B C
1 a b c
2 1 2 3
To read all worksheets we need a convenient function
read_excel_allsheets <- function(filename) {
sheets <- readxl::excel_sheets(filename)
sheets <- sheets[-1] # Skip sheet 1
x <- lapply(sheets, function(X) readxl::read_excel(filename, sheet = X, col_types = "numeric"))
names(x) <- sheets
x
}
dcfile <- "table0.77_dC_biospear.xlsx"
dc <- read_excel_allsheets(dcfile)
# Each component (eg dc[[1]]) is a tibble.
readr
Note: readr package is not designed to read Excel files.
Compared to base equivalents like read.csv(), readr is much faster and gives more convenient output: it never converts strings to factors, can parse date/times, and it doesn’t munge the column names.
1.0.0 released.
The read_csv() function from the readr package is as fast as fread() function from data.table package. For files beyond 100MB in size fread() and read_csv() can be expected to be around 5 times faster than read.csv(). See 5.3 of Efficient R Programming book.
Note that fread() can read-n a selection of the columns.
ggplot2
Books
- R for Data Science Chapter 28 Graphics for communication
- R Graphics Cookbook by Winston Chang. Lots of recipes. For example, the Axes chapter talks how to set/hide tick marks.
- The Hitchhiker's Guide to Ggplot2 in R
- ggplot2 book and its source code. Before I build the (pdf version) of the book, I need to follow this suggestion by running the following in R before calling make.
- Data Visualization for Social Science
devtools::install_github("hadley/oldbookdown")
- R Graph Essentials Essentials by David Lillis. Chapters 3 and 4.
Some examples:
- Top 50 ggplot2 Visualizations - The Master List
- http://blog.diegovalle.net/2015/01/the-74-most-violent-cities-in-mexico.html
- R Graph Catalog
Introduction
Cheat sheet
Examples from 'R for Data Science' book - Aesthetic mappings
ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy)) # template ggplot(data = <DATA>) + <GEOM_FUNCTION>(mapping = aes(<MAPPINGS>)) # add another variable through color, size, alpha or shape ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy, color = class)) ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy, size = class)) ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy, alpha = class)) ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy, shape = class)) ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy), color = "blue") # add another variable through facets ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy)) + facet_wrap(~ class, nrow = 2) # add another 2 variables through facets ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy)) + facet_grid(drv ~ cyl)
Examples from 'R for Data Science' book - Geometric objects
# Points ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy)) # Smoothed ggplot(data = mpg) + geom_smooth(mapping = aes(x = displ, y = hwy)) # Points + smoother ggplot(data = mpg) + geom_point(mapping = aes(x = displ, y = hwy)) + geom_smooth(mapping = aes(x = displ, y = hwy)) # Colored points + smoother ggplot(data = mpg, mapping = aes(x = displ, y = hwy)) + geom_point(mapping = aes(color = class)) + geom_smooth()
Examples from 'R for Data Science' book - Transformation
# y axis = counts # bar plot ggplot(data = diamonds) + geom_bar(mapping = aes(x = cut)) # Or ggplot(data = diamonds) + stat_count(mapping = aes(x = cut)) # y axis = proportion ggplot(data = diamonds) + geom_bar(mapping = aes(x = cut, y = ..prop.., group = 1)) # bar plot with 2 variables ggplot(data = diamonds) + geom_bar(mapping = aes(x = cut, fill = clarity))
ggthemr: Themes for ggplot2
ggedit & ggplotgui – interactive ggplot aesthetic and theme editor
- https://www.r-statistics.com/2016/11/ggedit-interactive-ggplot-aesthetic-and-theme-editor/
- https://github.com/gertstulp/ggplotgui/. It allows to change text (axis, title, font size), themes, legend, et al. A docker website was set up for the online version.
ggconf: Simpler Appearance Modification of 'ggplot2'
https://github.com/caprice-j/ggconf
Plotting individual observations and group means
https://drsimonj.svbtle.com/plotting-individual-observations-and-group-means-with-ggplot2
Colors
- How to expand color palette with ggplot and RColorBrewer
- palette_explorer() function from the tmaptools package. See selecting color palettes with shiny.
- Cookbook for R
- Sequential, diverging and qualitative colour scales/palettes from colorbrewer.org: scale_colour_brewer(), scale_fill_brewer(), ...
- http://colorbrewer2.org/
- It seems there is no choice of getting only 2 colors no matter which set name we can use
- To see the set names used in brewer.pal, see
- RColorBrewer::display.brewer.all()
- For example, Set1 from http://colorbrewer2.org/
- To list all R color names, colors()
- convert hex value to color names
library(plotrix) sapply(rainbow(4), color.id) sapply(RColorBrewer::brewer.pal(4, "Set1"), color.id)
Below is an example using the option scale_fill_brewer(palette = "Paired"). See the source code at gist. Note that only 'set1' and 'set3' palettes in qualitative scheme can support up to 12 classes.
According to the information from the colorbrew website, qualitative schemes do not imply magnitude differences between legend classes, and hues are used to create the primary visual differences between classes.
subplot
https://ikashnitsky.github.io/2017/subplots-in-maps/
Easy way to mix multiple graphs on the same page
- http://www.cookbook-r.com/Graphs/Multiple_graphs_on_one_page_(ggplot2)/
- Easy Way to Mix Multiple Graphs on The Same Page. Four packages are included: ggpubr, cowplot, gridExtra and grid.
- egg: Extensions for 'ggplot2', to Align Plots, Plot insets, and Set Panel Sizes.
- Why you should master small multiple chart
- gridExtra
x and y labels
https://stackoverflow.com/questions/10438752/adding-x-and-y-axis-labels-in-ggplot2 or the Labels part of the cheatsheet
You can set the labels with xlab() and ylab(), or make it part of the scale_*.* call.
labs(x = "sample size", y = "ngenes (glmnet)")
Legend title
scale_colour_manual("Treatment", values = c("black", "red"))
ylim and xlim in ggplot2
https://stackoverflow.com/questions/3606697/how-to-set-limits-for-axes-in-ggplot2-r-plots or the Zooming part of the cheatsheet
Use one of the following
- + scale_x_continuous(limits = c(-5000, 5000))
- + coord_cartesian(xlim = c(-5000, 5000))
- + xlim(-5000, 5000)
Center title
See the Legends part of the cheatsheet.
ggtitle("MY TITLE") +
theme(plot.title = element_text(hjust = 0.5))
Time series plot
- How to make a line chart with ggplot2
- Colour palettes. Note some palette options like Accent from the Qualitative category will give a warning message In RColorBrewer::brewer.pal(n, pal) : n too large, allowed maximum for palette Accent is 8.
Multiple lines plot https://stackoverflow.com/questions/14860078/plot-multiple-lines-data-series-each-with-unique-color-in-r
set.seed(45)
nc <- 9
df <- data.frame(x=rep(1:5, nc), val=sample(1:100, 5*nc),
variable=rep(paste0("category", 1:nc), each=5))
# plot
# http://colorbrewer2.org/#type=qualitative&scheme=Paired&n=9
ggplot(data = df, aes(x=x, y=val)) +
geom_line(aes(colour=variable)) +
scale_colour_manual(values=c("#a6cee3", "#1f78b4", "#b2df8a", "#33a02c", "#fb9a99", "#e31a1c", "#fdbf6f", "#ff7f00", "#cab2d6"))
Versus old fashion
dat <- matrix(runif(40,1,20),ncol=4) # make data
matplot(dat, type = c("b"),pch=1,col = 1:4) #plot
legend("topleft", legend = 1:4, col=1:4, pch=1) # optional legend
Github style calendar plot
- https://mvuorre.github.io/post/2016/2016-03-24-github-waffle-plot/
- https://gist.github.com/marcusvolz/84d69befef8b912a3781478836db9a75 from Create artistic visualisations with your exercise data
geom_errorbar(): error bars
- Can ggplot2 do this? https://www.nature.com/articles/nature25173/figures/1
- plotCI() from the plotrix package or geom_errorbar() from ggplot2 package
- http://sape.inf.usi.ch/quick-reference/ggplot2/geom_errorbar
- Vertical error bars
- Horizontal error bars
- Horizontal panel plot example and more
- R does not draw error bars out of the box. R has arrows() to create the error bars. Using just arrows(x0, y0, x1, y1, code=3, angle=90, length=.05, col). See
- Building Barplots with Error Bars. Note that the segments() statement is not necessary.
- https://www.rdocumentation.org/packages/graphics/versions/3.4.3/topics/arrows
- Toy example (see this nature paper)
set.seed(301)
x <- rnorm(10)
SE <- rnorm(10)
y <- 1:10
par(mfrow=c(2,1))
par(mar=c(0,4,4,4))
xlim <- c(-4, 4)
plot(x[1:5], 1:5, xlim=xlim, ylim=c(0+.1,6-.1), yaxs="i", xaxt = "n", ylab = "", pch = 16, las=1)
mtext("group 1", 4, las = 1, adj = 0, line = 1) # las=text rotation, adj=alignment, line=spacing
par(mar=c(5,4,0,4))
plot(x[6:10], 6:10, xlim=xlim, ylim=c(5+.1,11-.1), yaxs="i", ylab ="", pch = 16, las=1, xlab="")
arrows(x[6:10]-SE[6:10], 6:10, x[6:10]+SE[6:10], 6:10, code=3, angle=90, length=0)
mtext("group 2", 4, las = 1, adj = 0, line = 1)
text labels on scatterplots: ggrepel package
ggrepel package. Found on Some datasets for teaching data science by Rafael Irizarry.
graphics::smoothScatter
Data Manipulation & Tidyverse
Import
|
| readr, readxl
| haven, DBI, httr +----- Visualize ------+
| | ggplot2, ggvis |
| | |
Tidy ------------- Transform
tibble dplyr Model
tidyr | broom
+------ Model ---------+
- R for Data Science and tidyverse package (it is a collection of ggplot2, tibble, tidyr, readr, purrr & dplyr packages).
- tidyverse, among others, was used at Mining CRAN DESCRIPTION Files (tbl_df(), %>%, summarise(), count(), mutate(), arrange(), unite(), ggplot(), filter(), select(), ...). Note that there is a problem to reproduce the result. I need to run cran <- cran[, -14] to remove the MD5sum column.
- Compile R for Data Science to a PDF
- Data Wrangling with dplyr and tidyr Cheat Sheet
- Data Wrangling with Tidyverse from the Harvard Chan School of Public Health.
- Best packages for data manipulation in R. It demonstrates to perform the same tasks using data.table and dplyr packages. data.table is faster and it may be a go-to package when performance and memory are the constraints.
5 most useful data manipulation functions
- subset() for making subsets of data (natch)
- merge() for combining data sets in a smart and easy way
- melt()-reshape2 package for converting from wide to long data formats. See an example here where we want to combine multiple columns of values into 1 column.
- dcast()-reshape2 package for converting from long to wide data formats (or just use tapply()), and for making summary tables
- ddply()-plyr package for doing split-apply-combine operations, which covers a huge swath of the most tricky data operations
data.table
Fast aggregation of large data (e.g. 100GB in RAM or just several GB size file), fast ordered joins, fast add/modify/delete of columns by group using no copies at all, list columns and a fast file reader (fread).
OpenMP enabled compiler for Mac. This instruction works on my Mac El Capitan (10.11.6) when I need to upgrade the data.table version from 1.11.4 to 1.11.6.
Question: how to make use multicore with data.table package?
library(data.table)
x <- fread("mylargefile.txt")
- Note that 'x[, 2] always return 2. If you want to do the thing you want, use x[, 2, with=FALSE] or x[, V2] where V2 is the header name. See the FAQ #1 in data.table.
- Understanding data.table Rolling Joins
- Intro to The data.table Package
- In the Introduction to data.table vignette, the data.table::order() function is SLOWER than base::order() from my Odroid xu4 (running Ubuntu 14.04.4 trusty on uSD)
odt = data.table(col=sample(1e7)) (t1 <- system.time(ans1 <- odt[base::order(col)])) ## uses order from base R # user system elapsed # 2.730 0.210 2.947 (t2 <- system.time(ans2 <- odt[order(col)])) ## uses data.table's order # user system elapsed # 2.830 0.215 3.052 (identical(ans1, ans2)) # [1] TRUE
reshape & reshape2
- Data Shape Transformation With Reshape()
- Use acast() function in reshape2 package. It will convert data.frame used for analysis to a table-like data.frame good for display.
- http://lamages.blogspot.com/2013/10/creating-matrix-from-long-dataframe.html
tidyr
An evolution of reshape2. It's designed specifically for data tidying (not general reshaping or aggregating) and works well with dplyr data pipelines.
- vignette("tidy-data") & Cheat sheet
- Main functions
- Reshape data: gather() & spread()
- Split cells: separate() & unite()
- Handle missing: drop_na() & fill() & replace_na()
- http://blog.rstudio.org/2014/07/22/introducing-tidyr/
- http://rpubs.com/seandavi/GEOMetadbSurvey2014
- http://timelyportfolio.github.io/rCharts_factor_analytics/factors_with_new_R.html
- tidyr vs reshape2
Make wide tables long with gather() (see 6.3.1 of Efficient R Programming)
library(tidyr) library(efficient) data(pew) # wide table dim(pew) # 18 x 10, (religion, '<$10k', '$10--20k', '$20--30k', ..., '>150k') pewt <- gather(data = pew, key = Income, value = Count, -religion) dim(pew) # 162 x 3, (religion, Income, Count) args(gather) # function(data, key, value, ..., na.rm = FALSE, convert = FALSE, factor_key = FALSE)
where the three arguments of gather() requires:
- data: a data frame in which column names will become row vaues
- key: the name of the categorical variable into which the column names in the original datasets are converted.
- value: the name of cell value columns
In this example, the 'religion' column will not be included (-religion).
dplyr, plyr packages
- Essential functions: 3 rows functions, 3 column functions and 1 mixed function.
select, mutate, rename
+------------------+
filter + +
arrange + +
group_by + +
+ summarise +
+------------------+
- These functions works on data frames and tibble objects.
iris %>% filter(Species == "setosa") %>% count() head(iris %>% filter(Species == "setosa") %>% arrange(Sepal.Length))
- Data Transformation in the book R for Data Science. Five key functions in the dplyr package:
- Filter rows: filter()
- Arrange rows: arrange()
- Select columns: select()
- Add new variables: mutate()
- Grouped summaries: group_by() & summarise()
# filter
jan1 <- filter(flights, month == 1, day == 1)
filter(flights, month == 11 | month == 12)
filter(flights, arr_delay <= 120, dep_delay <= 120)
df <- tibble(x = c(1, NA, 3))
filter(df, x > 1)
filter(df, is.na(x) | x > 1)
# arrange
arrange(flights, year, month, day)
arrange(flights, desc(arr_delay))
# select
select(flights, year, month, day)
select(flights, year:day)
select(flights, -(year:day))
# mutate
flights_sml <- select(flights,
year:day,
ends_with("delay"),
distance,
air_time
)
mutate(flights_sml,
gain = arr_delay - dep_delay,
speed = distance / air_time * 60
)
# if you only want to keep the new variables
transmute(flights,
gain = arr_delay - dep_delay,
hours = air_time / 60,
gain_per_hour = gain / hours
)
# summarise()
by_day <- group_by(flights, year, month, day)
summarise(by_day, delay = mean(dep_delay, na.rm = TRUE))
# pipe. Note summarise() can return more than 1 variable.
delays <- flights %>%
group_by(dest) %>%
summarise(
count = n(),
dist = mean(distance, na.rm = TRUE),
delay = mean(arr_delay, na.rm = TRUE)
) %>%
filter(count > 20, dest != "HNL")
flights %>%
group_by(year, month, day) %>%
summarise(mean = mean(dep_delay, na.rm = TRUE))
- Videos
- Hands-on dplyr tutorial for faster data manipulation in R by Data School. At time 17:00, it compares the %>% operator, with() and aggregate() for finding group mean.
- https://youtu.be/aywFompr1F4 (shorter video) by Roger Peng
- https://youtu.be/8SGif63VW6E by Hadley Wickham
- Tidy eval: Programming with dplyr, tidyr, and ggplot2. Bang bang "!!" operator was introduced for use in a function call.
- Efficient R Programming
- Data wrangling: Transformation from R-exercises.
- Express Intro to dplyr by rollingyours.
- the dot.
- stringr and plyr A data.frame is pretty much a list of vectors, so we use plyr to apply over the list and stringr to search and replace in the vectors.
- https://randomjohn.github.io/r-maps-with-census-data/ dplyr and stringr are used
- 5 interesting subtle insights from TED videos data analysis in R
- What is tidy eval and why should I care?
stringr
- https://www.rstudio.com/wp-content/uploads/2016/09/RegExCheatsheet.pdf
- stringr Cheat sheet (2 pages, this will immediately download the pdf file)
magrittr
Instead of nested statements, it is using pipe operator %>%. So the code is easier to read. Impressive!
x %>% f # f(x) x %>% f(y) # f(x, y) x %>% f(arg=y) # f(x, arg=y) x %>% f(z, .) # f(z, x) x %>% f(y) %>% g(z) # g(f(x, y), z) x %>% select(which(colSums(!is.na(.))>0)) # remove columns with all missing data x %>% select(which(colSums(!is.na(.))>0)) %>% filter((rowSums(!is.na(.))>0)) # remove all-NA columns _and_ rows
suppressPackageStartupMessages(library("dplyr"))
starwars %>%
filter(., height > 200) %>%
select(., height, mass) %>%
head(.)
# instead of
starwars %>%
filter(height > 200) %>%
select(height, mass) %>%
head
iris$Species
iris[["Species"]]
iris %>%
`[[`("Species")
iris %>%
`[[`(5)
iris %>%
subset(select = "Species")
- Split-apply-combine: group + summarize + sort/arrange + top n. The following example is from Efficient R programming.
data(wb_ineq, package = "efficient")
wb_ineq %>%
filter(grepl("g", Country)) %>%
group_by(Year) %>%
summarise(gini = mean(gini, na.rm = TRUE)) %>%
arrange(desc(gini)) %>%
top_n(n = 5)
- Writing Pipe-friendly Functions
- http://rud.is/b/2015/02/04/a-step-to-the-right-in-r-assignments/
- http://rpubs.com/tjmahr/pipelines_2015
- http://danielmarcelino.com/i-loved-this-crosstable/
- http://moderndata.plot.ly/using-the-pipe-operator-in-r-with-plotly/
- Videos
# Examples from R for Data Science-Import, Tidy, Transform, Visualize, and Model
diamonds <- ggplot2::diamonds
diamonds2 <- diamonds %>% dplyr::mutate(price_per_carat = price / carat)
pryr::object_size(diamonds)
pryr::object_size(diamonds2)
pryr::object_size(diamonds, diamonds2)
rnorm(100) %>% matrix(ncol = 2) %>% plot() %>% str()
rnorm(100) %>% matrix(ncol = 2) %T>% plot() %>% str() # 'tee' pipe
# %T>% works like %>% except that it returns the lefthand side (rnorm(100) %>% matrix(ncol = 2))
# instead of the righthand side.
# If a function does not have a data frame based api, you can use %$%.
# It explodes out the variables in a data frame.
mtcars %$% cor(disp, mpg)
# For assignment, magrittr provides the %<>% operator
mtcars <- mtcars %>% transform(cyl = cyl * 2) # can be simplified by
mtcars %<>% transform(cyl = cyl * 2)
Upsides of using magrittr: no need to create intermediate objects, code is easy to read.
When not to use the pipe
- your pipes are longer than (say) 10 steps
- you have multiple inputs or outputs
- Functions that use the current environment: assign(), get(), load()
- Functions that use lazy evaluation: tryCatch(), try()
outer()
Genomic sequence
- chartr
> yourSeq <- "AAAACCCGGGTTTNNN"
> chartr("ACGT", "TGCA", yourSeq)
[1] "TTTTGGGCCCAAANNN"
lobstr package - dig into the internal representation and structure of R objects
Data Science
How to prepare data for collaboration
How to share data for collaboration. Especially Page 7 has some (raw data) variable coding guidelines.
- naming variables: using meaning variable names, no spacing in column header, avoiding separator (except an underscore)
- coding variables: be consistent, no spelling error
- date and time: YYYY-MM-DD (ISO 8601 standard). A gene symbol "Oct-4" will be interpreted as a date and reformatted in Excel.
- missing data: "NA". Not leave any cells blank.
- using a code book file (*.docx for example): any lengthy explanation about variables should be put here. See p5 for an example.
Five types of data:
- continuous
- oridinal
- categorical
- missing
- censored
Some extra from Data organization in spreadsheets (the paper appears in American Statistician)
- No empty cells
- Put one thing in a cell
- Make a rectangle
- No calculation in the raw data files
- Create a data dictionary (same as code book)
complete.cases()
Count the number of rows in a data frame that have missing values with
sum(!complete.cases(dF))
> tmp <- matrix(1:6, 3, 2)
> tmp
[,1] [,2]
[1,] 1 4
[2,] 2 5
[3,] 3 6
> tmp[2,1] <- NA
> complete.cases(tmp)
[1] TRUE FALSE TRUE
Wrangling categorical data in R
https://peerj.com/preprints/3163.pdf
Some approaches:
- options(stringAsFactors=FALSE)
- Use the tidyverse package
Base R approach:
GSS <- read.csv("XXX.csv")
GSS$BaseLaborStatus <- GSS$LaborStatus
levels(GSS$BaseLaborStatus)
summary(GSS$BaseLaborStatus)
GSS$BaseLaborStatus <- as.character(GSS$BaseLaborStatus)
GSS$BaseLaborStatus[GSS$BaseLaborStatus == "Temp not working"] <- "Temporarily not working"
GSS$BaseLaborStatus[GSS$BaseLaborStatus == "Unempl, laid off"] <- "Unemployed, laid off"
GSS$BaseLaborStatus[GSS$BaseLaborStatus == "Working fulltime"] <- "Working full time"
GSS$BaseLaborStatus[GSS$BaseLaborStatus == "Working parttime"] <- "Working part time"
GSS$BaseLaborStatus <- factor(GSS$BaseLaborStatus)
Tidyverse approach:
GSS <- GSS %>%
mutate(tidyLaborStatus =
recode(LaborStatus,
`Temp not working` = "Temporarily not working",
`Unempl, laid off` = "Unemployed, laid off",
`Working fulltime` = "Working full time",
`Working parttime ` = "Working part time"))
jpeg
If we want to create the image on this wiki left hand side panel, we can use the jpeg package to read an existing plot and then edit and save it.
We can also use the jpeg package to import and manipulate a jpg image. See Fun with Heatmaps and Plotly.
Cairo
See White strips problem in png() or tiff().
cairoDevice
PS. Not sure the advantage of functions in this package compared to R's functions (eg. Cairo_svg() vs svg()).
For ubuntu OS, we need to install 2 libraries and 1 R package RGtk2.
sudo apt-get install libgtk2.0-dev libcairo2-dev
On Windows OS, we may got the error: unable to load shared object 'C:/Program Files/R/R-3.0.2/library/cairoDevice/libs/x64/cairoDevice.dll' . We need to follow the instruction in here.
igraph
creating directed networks with igraph
Identifying dependencies of R functions and scripts
https://stackoverflow.com/questions/8761857/identifying-dependencies-of-r-functions-and-scripts
library(mvbutils)
foodweb(where = "package:batr")
foodweb( find.funs("package:batr"), prune="survRiskPredict", lwd=2)
foodweb( find.funs("package:batr"), prune="classPredict", lwd=2)
iterators
Iterator is useful over for-loop if the data is already a collection. It can be used to iterate over a vector, data frame, matrix, file
Iterator can be combined to use with foreach package http://www.exegetic.biz/blog/2013/11/iterators-in-r/ has more elaboration.
Colors
- http://www.bauer.uh.edu/parks/truecolor.htm Interactive RGB, Alpha and Color Picker
- http://deanattali.com/blog/colourpicker-package/ Not sure what it is doing
- How to Choose the Best Colors For Your Data Charts
- How to expand color palette with ggplot and RColorBrewer
- Color names in R
colortools
Tools that allow users generate color schemes and palettes
colourpicker
A Colour Picker Tool for Shiny and for Selecting Colours in Plots
inlmisc
GetTolColors(). Lots of examples.
rex
Friendly Regular Expressions
formatR
The best strategy to avoid failure is to put comments in complete lines or after complete R expressions.
See also this discussion on stackoverflow talks about R code reformatting.
library(formatR)
tidy_source("Input.R", file = "output.R", width.cutoff=70)
tidy_source("clipboard")
# default width is getOption("width") which is 127 in my case.
Some issues
- Comments appearing at the beginning of a line within a long complete statement. This will break tidy_source().
cat("abcd",
# This is my comment
"defg")
will result in
> tidy_source("clipboard")
Error in base::parse(text = code, srcfile = NULL) :
3:1: unexpected string constant
2: invisible(".BeGiN_TiDy_IdEnTiFiEr_HaHaHa# This is my comment.HaHaHa_EnD_TiDy_IdEnTiFiEr")
3: "defg"
^
- Comments appearing at the end of a line within a long complete statement won't break tidy_source() but tidy_source() cannot re-locate/tidy the comma sign.
cat("abcd"
,"defg" # This is my comment
,"ghij")
will become
cat("abcd", "defg" # This is my comment
, "ghij")
Still bad!!
- Comments appearing at the end of a line within a long complete statement breaks tidy_source() function. For example,
cat("</p>",
"<HR SIZE=5 WIDTH=\"100%\" NOSHADE>",
ifelse(codeSurv == 0,"<h3><a name='Genes'><b><u>Genes which are differentially expressed among classes:</u></b></a></h3>", #4/9/09
"<h3><a name='Genes'><b><u>Genes significantly associated with survival:</u></b></a></h3>"),
file=ExternalFileName, sep="\n", append=T)
will result in
> tidy_source("clipboard", width.cutoff=70)
Error in base::parse(text = code, srcfile = NULL) :
3:129: unexpected SPECIAL
2: "<HR SIZE=5 WIDTH=\"100%\" NOSHADE>" ,
3: ifelse ( codeSurv == 0 , "<h3><a name='Genes'><b><u>Genes which are differentially expressed among classes:</u></b></a></h3>" , %InLiNe_IdEnTiFiEr%
- width.cutoff parameter is not always working. For example, there is no any change for the following snippet though I hope it will move the cat() to the next line.
if (codePF & !GlobalTest & !DoExactPermTest) cat(paste("Multivariate Permutations test was computed based on",
NumPermutations, "random permutations"), "<BR>", " ", file = ExternalFileName,
sep = "\n", append = T)
- It merges lines though I don't always want to do that. For example
cat("abcd"
,"defg"
,"ghij")
will become
cat("abcd", "defg", "ghij")
Download papers
biorxivr
Search and Download Papers from the bioRxiv Preprint Server
aRxiv
Interface to the arXiv API
pdftools
- http://ropensci.org/blog/2016/03/01/pdftools-and-jeroen
- http://r-posts.com/how-to-extract-data-from-a-pdf-file-with-r/
- https://ropensci.org/technotes/2018/12/14/pdftools-20/
aside: set it aside
An RStudio addin to run long R commands aside your current session.
Teaching
- smovie: Some Movies to Illustrate Concepts in Statistics
packrat for reproducible search
- CRAN & Github
- Videos:
- Packrat will not only store all packages, but also all project files.
- Packrat is integrated in RStudio’s user interface. It allows you to share projects along co-workers easily. See Using Packrat with RStudio.
- limitations.
- Git and packrat. The packrat/src directory can be very large. If you don't want them available in your git-repo, you simply add packrat/src/ to the .gitignore. But, this will mean that anyone accessing the git-repo will not have access to the package source code, and the files will be downloaded from CRAN, or from wherever the source line dictates within the packrat.lock file.
Create a snapshot:
- Do we really need to call packrat::snapshot()? The walk through page says it is not needed but the lock file is not updated from my testing.
- I got an error when it is trying to fetch the source code from bioconductor and local repositories: packrat is trying to fetch the source from CRAN in these two packages.
- On normal case, the packrat/packrat.lock file contains two entries in 'Repos' field (line 4).
- The cause of the error is I ran snapshot() after I quitted R and entered again. So the solution is to add bioc and local repositories to options(repos).
- So what is important of running snapshot()?
- Check out the forum.
> dir.create("~/projects/babynames", recu=T)
> packrat::init("~/projects/babynames")
Initializing packrat project in directory:
- "~/projects/babynames"
Adding these packages to packrat:
_
packrat 0.4.9-3
Fetching sources for packrat (0.4.9-3) ... OK (CRAN current)
Snapshot written to '/home/brb/projects/babynames/packrat/packrat.lock'
Installing packrat (0.4.9-3) ...
OK (built source)
Initialization complete!
Unloading packages in user library:
- packrat
Packrat mode on. Using library in directory:
- "~/projects/babynames/packrat/lib"
> install.packages("reshape2")
> packrat::snapshot()
> system("tree -L 2 ~/projects/babynames/packrat/")
/home/brb/projects/babynames/packrat/
├── init.R
├── lib
│ └── x86_64-pc-linux-gnu
├── lib-ext
│ └── x86_64-pc-linux-gnu
├── lib-R # base packages
│ └── x86_64-pc-linux-gnu
├── packrat.lock
├── packrat.opts
└── src
├── bitops
├── glue
├── magrittr
├── packrat
├── plyr
├── Rcpp
├── reshape2
├── stringi
└── stringr
Restoring snapshots:
Suppose a packrat project was created on Ubuntu 16.04 and we now want to repeat the analysis on Ubuntu 18.04. We first copy the whole project directory ('babynames') to Ubuntu 18.04. Then we should delete the library subdirectory ('packrat/lib') which contains binary files (*.so) that do not work on the new OS. After we delete the library subdirectory, start R from the project directory. Now if we run packrat::restore() command, it will re-install all missing libraries. Bingo! NOTE: Maybe I should use packrat::bundle() instead of manually copy the whole project folder.
Note: some OS level libraries (e.g. libXXX-dev) need to be installed manually beforehand in order for the magic to work.
$ rm -rf ~/projects/babynames/packrat/lib
$ cd ~/projects/babynames/
$ R
>
> packrat::status()
> remove.packages("plyr")
> packrat::status()
> packrat::restore()
Set Up a Custom CRAN-like Repository:
See https://rstudio.github.io/packrat/custom-repos.html. Note the personal repository name ('sushi' in this example) used in "Repository" field of the personal package will be used in <packrat/packrat.lock> file. So as long as we work on the same computer, it is easy to restore a packrat project containing packages coming from personal repository.
- packrat::init()
- packrat::snapshot(), packrat::restore()
- packrat::clean()
- packrat::status()
- packrat::install_local() # http://rstudio.github.io/packrat/limitations.html
- packrat::bundle() # see @28:44 of the video, packrat::unbundle() # see @29:17 of the same video. This will rebuild all packages
- packrat::on(), packrat::off()
- packrat::get_opts()
- packrat::set_opts() # http://rstudio.github.io/packrat/limitations.html
- packrat::opts$local.repos("~/local-cran")
- packrat::opts$external.packages(c("devtools")) # break the isolation
- packrat::extlib()
- packrat::with_extlib()
- packrat::project_dir(), .libPaths()
Warning
- If we download and modify some function definition from a package in CRAN without changing DESCRIPTION file or the package name, the snapshot created using packrat::snapshot() will contain the package source from CRAN instead of local repository. This is because (I guess) the DESCRIPTION file contains a field 'Repository' with the value 'CRAN'.
Text to speech
Text-to-Speech with the googleLanguageR package
Weather data
- prism package
- Weatherbase
logR
https://github.com/jangorecki/logR
Progress bar
https://github.com/r-lib/progress#readme
Configurable Progress bars, they may include percentage, elapsed time, and/or the estimated completion time. They work in terminals, in 'Emacs' 'ESS', 'RStudio', 'Windows' 'Rgui' and the 'macOS'.
Different ways of using R
dyn.load
Error: “not resolved from current namespace” error, when calling C routines from R
Solution: add getNativeSymbolInfo() around your C/Fortran symbols. Search Google:r dyn.load not resolved from current namespace
R call C/C++
Mainly talks about .C() and .Call().
- R-Extension manual of course.
- http://faculty.washington.edu/kenrice/sisg-adv/sisg-07.pdf
- http://www.stat.berkeley.edu/scf/paciorek-cppWorkshop.pdf (Very useful)
- http://www.stat.harvard.edu/ccr2005/
- http://mazamascience.com/WorkingWithData/?p=1099
SEXP
Some examples from packages
- sva package has one C code function
R call Fortran
- R call Fortran 90
- The Need for Speed Part 1: Building an R Package with Fortran (or C) (Very detailed)
Embedding R
- See Writing for R Extensions Manual Chapter 8.
- Talk by Simon Urbanek in UseR 2004.
- Technical report by Friedrich Leisch in 2007.
- https://stat.ethz.ch/pipermail/r-help/attachments/20110729/b7d86ed7/attachment.pl
An very simple example (do not return from shell) from Writing R Extensions manual
The command-line R front-end, R_HOME/bin/exec/R, is one such example. Its source code is in file <src/main/Rmain.c>.
This example can be run by
R_HOME/bin/R CMD R_HOME/bin/exec/R
Note:
- R_HOME/bin/exec/R is the R binary. However, it couldn't be launched directly unless R_HOME and LD_LIBRARY_PATH are set up. Again, this is explained in Writing R Extension manual.
- R_HOME/bin/R is a shell-script front-end where users can invoke it. It sets up the environment for the executable. It can be copied to /usr/local/bin/R. When we run R_HOME/bin/R, it actually runs R_HOME/bin/R CMD R_HOME/bin/exec/R (see line 259 of R_HOME/bin/R as in R 3.0.2) so we know the important role of R_HOME/bin/exec/R.
More examples of embedding can be found in tests/Embedding directory. Read <index.html> for more information about these test examples.
An example from Bioconductor workshop
- What is covered in this section is different from Create and use a standalone Rmath library.
- Use eval() function. See R-Ext 8.1 and 8.2 and 5.11.
- http://stackoverflow.com/questions/2463437/r-from-c-simplest-possible-helloworld (obtained from searching R_tryEval on google)
- http://stackoverflow.com/questions/7457635/calling-r-function-from-c
Example: Create <embed.c> file
#include <Rembedded.h>
#include <Rdefines.h>
static void doSplinesExample();
int
main(int argc, char *argv[])
{
Rf_initEmbeddedR(argc, argv);
doSplinesExample();
Rf_endEmbeddedR(0);
return 0;
}
static void
doSplinesExample()
{
SEXP e, result;
int errorOccurred;
// create and evaluate 'library(splines)'
PROTECT(e = lang2(install("library"), mkString("splines")));
R_tryEval(e, R_GlobalEnv, &errorOccurred);
if (errorOccurred) {
// handle error
}
UNPROTECT(1);
// 'options(FALSE)' ...
PROTECT(e = lang2(install("options"), ScalarLogical(0)));
// ... modified to 'options(example.ask=FALSE)' (this is obscure)
SET_TAG(CDR(e), install("example.ask"));
R_tryEval(e, R_GlobalEnv, NULL);
UNPROTECT(1);
// 'example("ns")'
PROTECT(e = lang2(install("example"), mkString("ns")));
R_tryEval(e, R_GlobalEnv, &errorOccurred);
UNPROTECT(1);
}
Then build the executable. Note that I don't need to create R_HOME variable.
cd tar xzvf cd R-3.0.1 ./configure --enable-R-shlib make cd tests/Embedding make ~/R-3.0.1/bin/R CMD ./Rtest nano embed.c # Using a single line will give an error and cannot not show the real problem. # ../../bin/R CMD gcc -I../../include -L../../lib -lR embed.c # A better way is to run compile and link separately gcc -I../../include -c embed.c gcc -o embed embed.o -L../../lib -lR -lRblas ../../bin/R CMD ./embed
Note that if we want to call the executable file ./embed directly, we shall set up R environment by specifying R_HOME variable and including the directories used in linking R in LD_LIBRARY_PATH. This is based on the inform provided by Writing R Extensions.
export R_HOME=/home/brb/Downloads/R-3.0.2 export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/home/brb/Downloads/R-3.0.2/lib ./embed # No need to include R CMD in front.
Question: Create a data frame in C? Answer: Use data.frame() via an eval() call from C. Or see the code is stats/src/model.c, as part of model.frame.default. Or using Rcpp as here.
Reference http://bioconductor.org/help/course-materials/2012/Seattle-Oct-2012/AdvancedR.pdf
Create a Simple Socket Server in R
This example is coming from this paper.
Create an R function
simpleServer <- function(port=6543)
{
sock <- socketConnection ( port=port , server=TRUE)
on.exit(close( sock ))
cat("\nWelcome to R!\nR>" ,file=sock )
while(( line <- readLines ( sock , n=1)) != "quit")
{
cat(paste("socket >" , line , "\n"))
out<- capture.output (try(eval(parse(text=line ))))
writeLines ( out , con=sock )
cat("\nR> " ,file =sock )
}
}
Then run simpleServer(). Open another terminal and try to communicate with the server
$ telnet localhost 6543 Trying 127.0.0.1... Connected to localhost. Escape character is '^]'. Welcome to R! R> summary(iris[, 3:5]) Petal.Length Petal.Width Species Min. :1.000 Min. :0.100 setosa :50 1st Qu.:1.600 1st Qu.:0.300 versicolor:50 Median :4.350 Median :1.300 virginica :50 Mean :3.758 Mean :1.199 3rd Qu.:5.100 3rd Qu.:1.800 Max. :6.900 Max. :2.500 R> quit Connection closed by foreign host.
Rserve
Note the way of launching Rserve is like the way we launch C program when R was embedded in C. See Call R from C/C++ or Example from Bioconductor workshop.
See my Rserve page.
(Commercial) StatconnDcom
R.NET
rJava
- A primer in using Java from R - part 1
- Note rJava is needed by xlsx package.
Terminal
# jdk 7 sudo apt-get install openjdk-7-* update-alternatives --config java # oracle jdk 8 sudo add-apt-repository -y ppa:webupd8team/java sudo apt-get update echo debconf shared/accepted-oracle-license-v1-1 select true | sudo debconf-set-selections echo debconf shared/accepted-oracle-license-v1-1 seen true | sudo debconf-set-selections sudo apt-get -y install openjdk-8-jdk
and then run the following (thanks to http://stackoverflow.com/questions/12872699/error-unable-to-load-installed-packages-just-now) to fix an error: libjvm.so: cannot open shared object file: No such file or directory.
- Create the file /etc/ld.so.conf.d/java.conf with the following entries:
/usr/lib/jvm/java-8-oracle/jre/lib/amd64 /usr/lib/jvm/java-8-oracle/jre/lib/amd64/server
- And then run sudo ldconfig
Now go back to R
install.packages("rJava")
Done!
If above does not work, a simple way is by (under Ubuntu) running
sudo apt-get install r-cran-rjava
which will create new package 'default-jre' (under /usr/lib/jvm) and 'default-jre-headless'.
RCaller
RApache
littler
Provides hash-bang (#!) capability for R
Difference between Rscript and littler and Whats the difference between Rscript and R CMD BATCH
We can install littler using two ways.
- install.packages("littler"). This will install the latest version but the binary 'r' program is only available under the package/bin directory (eg ~/R/x86_64-pc-linux-gnu-library/3.4/littler/bin/r). You need to create a soft link in order to access it globally.
- sudo apt install littler. This will install 'r' globally; however, the installed version may be old.
After the installation, vignette contains several examples. The off-line vignette has a table of contents. Nice! The web version of examples does not have the TOC.
r was not meant to run interactively like R. See man r.
RInside: Embed R in C++
See RInside
(From RInside documentation) The RInside package makes it easier to embed R in your C++ applications. There is no code you would execute directly from the R environment. Rather, you write C++ programs that embed R which is illustrated by some the included examples.
The included examples are armadillo, eigen, mpi, qt, standard, threads and wt.
To run 'make' when we don't have a global R, we should modify the file <Makefile>. Also if we just want to create one executable file, we can do, for example, 'make rinside_sample1'.
To run any executable program, we need to specify LD_LIBRARY_PATH variable, something like
export LD_LIBRARY_PATH=$LD_LIBRARY_PATH:/home/brb/Downloads/R-3.0.2/lib
The real build process looks like (check <Makefile> for completeness)
g++ -I/home/brb/Downloads/R-3.0.2/include \
-I/home/brb/Downloads/R-3.0.2/library/Rcpp/include \
-I/home/brb/Downloads/R-3.0.2/library/RInside/include -g -O2 -Wall \
-I/usr/local/include \
rinside_sample0.cpp \
-L/home/brb/Downloads/R-3.0.2/lib -lR -lRblas -lRlapack \
-L/home/brb/Downloads/R-3.0.2/library/Rcpp/lib -lRcpp \
-Wl,-rpath,/home/brb/Downloads/R-3.0.2/library/Rcpp/lib \
-L/home/brb/Downloads/R-3.0.2/library/RInside/lib -lRInside \
-Wl,-rpath,/home/brb/Downloads/R-3.0.2/library/RInside/lib \
-o rinside_sample0
Hello World example of embedding R in C++.
#include <RInside.h> // for the embedded R via RInside
int main(int argc, char *argv[]) {
RInside R(argc, argv); // create an embedded R instance
R["txt"] = "Hello, world!\n"; // assign a char* (string) to 'txt'
R.parseEvalQ("cat(txt)"); // eval the init string, ignoring any returns
exit(0);
}
The above can be compared to the Hello world example in Qt.
#include <QApplication.h>
#include <QPushButton.h>
int main( int argc, char **argv )
{
QApplication app( argc, argv );
QPushButton hello( "Hello world!", 0 );
hello.resize( 100, 30 );
app.setMainWidget( &hello );
hello.show();
return app.exec();
}
RFortran
RFortran is an open source project with the following aim:
To provide an easy to use Fortran software library that enables Fortran programs to transfer data and commands to and from R.
It works only on Windows platform with Microsoft Visual Studio installed:(
Call R from other languages
JRI
ryp2
http://rpy.sourceforge.net/rpy2.html
Create a standalone Rmath library
R has many math and statistical functions. We can easily use these functions in our C/C++/Fortran. The definite guide of doing this is on Chapter 9 "The standalone Rmath library" of R-admin manual.
Here is my experience based on R 3.0.2 on Windows OS.
Create a static library <libRmath.a> and a dynamic library <Rmath.dll>
Suppose we have downloaded R source code and build R from its source. See Build_R_from_its_source. Then the following 2 lines will generate files <libRmath.a> and <Rmath.dll> under C:\R\R-3.0.2\src\nmath\standalone directory.
cd C:\R\R-3.0.2\src\nmath\standalone make -f Makefile.win
Use Rmath library in our code
set CPLUS_INCLUDE_PATH=C:\R\R-3.0.2\src\include set LIBRARY_PATH=C:\R\R-3.0.2\src\nmath\standalone # It is not LD_LIBRARY_PATH in above. # Created <RmathEx1.cpp> from the book "Statistical Computing in C++ and R" web site # http://math.la.asu.edu/~eubank/CandR/ch4Code.cpp # It is OK to save the cpp file under any directory. # Force to link against the static library <libRmath.a> g++ RmathEx1.cpp -lRmath -lm -o RmathEx1.exe # OR g++ RmathEx1.cpp -Wl,-Bstatic -lRmath -lm -o RmathEx1.exe # Force to link against dynamic library <Rmath.dll> g++ RmathEx1.cpp Rmath.dll -lm -o RmathEx1Dll.exe
Test the executable program. Note that the executable program RmathEx1.exe can be transferred to and run in another computer without R installed. Isn't it cool!
c:\R>RmathEx1 Enter a argument for the normal cdf: 1 Enter a argument for the chi-squared cdf: 1 Prob(Z <= 1) = 0.841345 Prob(Chi^2 <= 1)= 0.682689
Below is the cpp program <RmathEx1.cpp>.
//RmathEx1.cpp
#define MATHLIB_STANDALONE
#include <iostream>
#include "Rmath.h"
using std::cout; using std::cin; using std::endl;
int main()
{
double x1, x2;
cout << "Enter a argument for the normal cdf:" << endl;
cin >> x1;
cout << "Enter a argument for the chi-squared cdf:" << endl;
cin >> x2;
cout << "Prob(Z <= " << x1 << ") = " <<
pnorm(x1, 0, 1, 1, 0) << endl;
cout << "Prob(Chi^2 <= " << x2 << ")= " <<
pchisq(x2, 1, 1, 0) << endl;
return 0;
}
Calling R.dll directly
See Chapter 8.2.2 of R Extensions. This is related to embedding R under Windows. The file <R.dll> on Windows is like <libR.so> on Linux.
bookdown.org
The website is full of open-source books written with R markdown.
- Announce bookdown
- bookdown package: Authoring Books and Technical Documents with R Markdown
- Compile R for Data Science to a PDF
Writing a R book and self-publishing it in Amazon
Publish R results
5 amazing free tools that can help with publishing R results and blogging
Scheduling R Markdown Reports via Email
http://www.analyticsforfun.com/2016/01/scheduling-r-markdown-reports-via-email.html
Create presentation file (beamer)
- http://rmarkdown.rstudio.com/beamer_presentation_format.html
- http://www.theresearchkitchen.com/archives/1017 (markdown and presentation files)
- http://rmarkdown.rstudio.com/
- Create Rmd file first in Rstudio by File -> R markdown. Select Presentation > choose pdf (beamer) as output format.
- Edit the template created by RStudio.
- Click 'Knit pdf' button (Ctrl+Shift+k) to create/display the pdf file.
An example of Rmd is
---
title: "My Example"
author: You Know Me
date: Dec 32, 2014
output: beamer_presentation
---
## R Markdown
This is an R Markdown presentation. Markdown is a simple formatting syntax for authoring HTML, PDF, and MS Word documents.
For more details on using R Markdown see <http://rmarkdown.rstudio.com>.
When you click the **Knit** button a document will be generated that includes both content as well as the output of any
embedded R code chunks within the document.
## Slide with Bullets
- Bullet 1
- Bullet 2
- Bullet 3. Mean is $\frac{1}{n} \sum_{i=1}^n x_i$.
$$
\mu = \frac{1}{n} \sum_{i=1}^n x_i
$$
## New slide

## Slide with R Code and Output
```{r}
summary(cars)
```
## Slide with Plot
```{r, echo=FALSE}
plot(cars)
```
Create HTML report
ReportingTools (Jason Hackney) from Bioconductor.
htmlTable package
The htmlTable package is intended for generating tables using HTML formatting. This format is compatible with Markdown when used for HTML-output. The most basic table can easily be created by just passing a matrix or a data.frame to the htmlTable-function.
- http://cran.r-project.org/web/packages/htmlTable/vignettes/general.html
- http://gforge.se/2014/01/fast-track-publishing-using-knitr-part-iv/
formattable
- https://github.com/renkun-ken/formattable
- http://www.magesblog.com/2016/01/formatting-table-output-in-r.html
- Make Beautiful Tables with the Formattable Package
htmltab package
This package is NOT used to CREATE html report but EXTRACT html table.
ztable package
Makes zebra-striped tables (tables with alternating row colors) in LaTeX and HTML formats easily from a data.frame, matrix, lm, aov, anova, glm or coxph objects.
Create academic report
reports package in CRAN and in github repository. The youtube video gives an overview of the package.
Create pdf and epub files
# Idea:
# knitr pdflatex
# rnw -------> tex ----------> pdf
library(knitr)
knit("example.rnw") # create example.tex file
- A very simple example <002-minimal.Rnw> from yihui.name works fine on linux.
git clone https://github.com/yihui/knitr-examples.git
- <knitr-minimal.Rnw>. I have no problem to create pdf file on Windows but still cannot generate pdf on Linux from tex file. Some people suggested to run sudo apt-get install texlive-fonts-recommended to install missing fonts. It works!
To see a real example, check out DESeq2 package (inst/doc subdirectory). In addition to DESeq2, I also need to install DESeq, BiocStyle, airway, vsn, gplots, and pasilla packages from Bioconductor. Note that, it is best to use sudo/admin account to install packages.
Or starts with markdown file. Download the example <001-minimal.Rmd> and remove the last line of getting png file from internet.
# Idea:
# knitr pandoc
# rmd -------> md ----------> pdf
git clone https://github.com/yihui/knitr-examples.git
cd knitr-examples
R -e "library(knitr); knit('001-minimal.Rmd')"
pandoc 001-minimal.md -o 001-minimal.pdf # require pdflatex to be installed !!
To create an epub file (not success yet on Windows OS, missing figures on Linux OS)
# Idea:
# knitr pandoc
# rnw -------> tex ----------> markdown or epub
library(knitr)
knit("DESeq2.Rnw") # create DESeq2.tex
system("pandoc -f latex -t markdown -o DESeq2.md DESeq2.tex")
## Windows OS, epub cannot be built
pandoc:
Error:
"source" (line 41, column 7):
unexpected "k"
expecting "{document}"
## Linux OS, epub missing figures and R codes.
## First install texlive base and extra packages
## sudo apt-get install texlive-latex-base texlive-latex-extra
pandoc: Could not find media `figure/SchwederSpjotvoll-1', skipping...
pandoc: Could not find media `figure/sortedP-1', skipping...
pandoc: Could not find media `figure/figHeatmap2c-1', skipping...
pandoc: Could not find media `figure/figHeatmap2b-1', skipping...
pandoc: Could not find media `figure/figHeatmap2a-1', skipping...
pandoc: Could not find media `figure/plotCountsAdv-1', skipping...
pandoc: Could not find media `figure/plotCounts-1', skipping...
pandoc: Could not find media `figure/MA-1', skipping...
pandoc: Could not find media `figure/MANoPrior-1', skipping...
The problems are at least
- figures need to be generated under the same directory as the source code
- figures cannot be in the format of pdf (DESeq2 generates both pdf and png files format)
- missing R codes
Convert tex to epub
kable() for tables
Create Tables In LaTeX, HTML, Markdown And ReStructuredText
- https://rmarkdown.rstudio.com/lesson-7.html
- https://stackoverflow.com/questions/20942466/creating-good-kable-output-in-rstudio
- http://kbroman.org/knitr_knutshell/pages/figs_tables.html
- https://blogs.reed.edu/ed-tech/2015/10/creating-nice-tables-using-r-markdown/
- kableExtra package
Create Word report
knitr + pandoc
- http://www.r-statistics.com/2013/03/write-ms-word-document-using-r-with-as-little-overhead-as-possible/
- http://www.carlboettiger.info/2012/04/07/writing-reproducibly-in-the-open-with-knitr.html
- http://rmarkdown.rstudio.com/articles_docx.html
It is better to create rmd file in RStudio. Rstudio provides a template for rmd file and it also provides a quick reference to R markdown language.
# Idea:
# knitr pandoc
# rmd -------> md --------> docx
library(knitr)
knit2html("example.rmd") #Create md and html files
and then
FILE <- "example"
system(paste0("pandoc -o ", FILE, ".docx ", FILE, ".md"))
Note. For example reason, if I play around the above 2 commands for several times, the knit2html() does not work well. However, if I click 'Knit HTML' button on the RStudio, it then works again.
Another way is
library(pander) name = "demo" knit(paste0(name, ".Rmd"), encoding = "utf-8") Pandoc.brew(file = paste0(name, ".md"), output = paste0(-name, "docx"), convert = "docx")
Note that once we have used knitr command to create a md file, we can use pandoc shell command to convert it to different formats:
- A pdf file: pandoc -s report.md -t latex -o report.pdf
- A html file: pandoc -s report.md -o report.html (with the -c flag html files can be added easily)
- Openoffice: pandoc report.md -o report.odt
- Word docx: pandoc report.md -o report.docx
We can also create the epub file for reading on Kobo ereader. For example, download this file and save it as example.Rmd. I need to remove the line containing the link to http://i.imgur.com/RVNmr.jpg since it creates an error when I run pandoc (not sure if it is the pandoc version I have is too old). Now we just run these 2 lines to get the epub file. Amazing!
knit("example.Rmd")
pandoc("example.md", format="epub")
PS. If we don't remove the link, we will get an error message (pandoc 1.10.1 on Windows 7)
> pandoc("Rmd_to_Epub.md", format="epub")
executing pandoc -f markdown -t epub -o Rmd_to_Epub.epub "Rmd_to_Epub.utf8md"
pandoc.exe: .\.\http://i.imgur.com/RVNmr.jpg: openBinaryFile: invalid argument (Invalid argument)
Error in (function (input, format, ext, cfg) : conversion failed
In addition: Warning message:
running command 'pandoc -f markdown -t epub -o Rmd_to_Epub.epub "Rmd_to_Epub.utf8md"' had status 1
pander
Try pandoc[1] with a minimal reproducible example, you might give a try to my "pander" package [2] too:
library(pander)
Pandoc.brew(system.file('examples/minimal.brew', package='pander'),
output = tempfile(), convert = 'docx')
Where the content of the "minimal.brew" file is something you might have got used to with Sweave - although it's using "brew" syntax instead. See the examples of pander [3] for more details. Please note that pandoc should be installed first, which is pretty easy on Windows.
- http://johnmacfarlane.net/pandoc/
- http://rapporter.github.com/pander/
- http://rapporter.github.com/pander/#examples
R2wd
Use R2wd package. However, only 32-bit R is allowed and sometimes it can not produce all 'table's.
> library(R2wd)
> wdGet()
Loading required package: rcom
Loading required package: rscproxy
rcom requires a current version of statconnDCOM installed.
To install statconnDCOM type
installstatconnDCOM()
This will download and install the current version of statconnDCOM
You will need a working Internet connection
because installation needs to download a file.
Error in if (wdapp[["Documents"]][["Count"]] == 0) wdapp[["Documents"]]$Add() :
argument is of length zero
The solution is to launch 32-bit R instead of 64-bit R since statconnDCOM does not support 64-bit R.
Convert from pdf to word
The best rendering of advanced tables is done by converting from pdf to Word. See http://biostat.mc.vanderbilt.edu/wiki/Main/SweaveConvert
rtf
Use rtf package for Rich Text Format (RTF) Output.
xtable
Package xtable will produce html output.
print(xtable(X), type="html")
If you save the file and then open it with Word, you will get serviceable results. I've had better luck copying the output from xtable and pasting it into Excel.
ReporteRs
Microsoft Word, Microsoft Powerpoint and HTML documents generation from R. The source code is hosted on https://github.com/davidgohel/ReporteRs
R Graphs Gallery
- Romain François
- R Graph Catalog written using R + Shiny. The source code is available on Github.
- Forest plot. See the packages rmeta and forestplot. The forest plot can be used to plot the quantities like relative risk (with 95% CI) in survival data.
COM client or server
Client
RDCOMClient where excel.link depends on it.
Server
Use R under proxy
http://support.rstudio.org/help/kb/faq/configuring-r-to-use-an-http-proxy
RStudio
- Github
- Installing RStudio (1.0.44) on Ubuntu will not install Java even the source code contains 37.5% Java??
- Preview
rstudio.cloud
Launch RStudio
Create .Rproj file
If you have an existing package that doesn't have an .Rproj file, you can use devtools::use_rstudio("path/to/package") to add it.
With an RStudio project file, you can
- Restore .RData into workspace at startup
- Save workspace to .RData on exit
- Always save history (even if no saving .RData)
- etc
package search
https://github.com/RhoInc/CRANsearcher
Git
Visual Studio
R and Python support now built in to Visual Studio 2017
List files using regular expression
- Extension
list.files(pattern = "\\.txt$")
where the dot (.) is a metacharacter. It is used to refer to any character.
- Start with
list.files(pattern = "^Something")
Using Sys.glob()"' as
> Sys.glob("~/Downloads/*.txt")
[1] "/home/brb/Downloads/ip.txt" "/home/brb/Downloads/valgrind.txt"
Hidden tool: rsync in Rtools
c:\Rtools\bin>rsync -avz "/cygdrive/c/users/limingc/Downloads/a.exe" "/cygdrive/c/users/limingc/Documents/" sending incremental file list a.exe sent 323142 bytes received 31 bytes 646346.00 bytes/sec total size is 1198416 speedup is 3.71 c:\Rtools\bin>
And rsync works best when we need to sync folder.
c:\Rtools\bin>rsync -avz "/cygdrive/c/users/limingc/Downloads/binary" "/cygdrive/c/users/limingc/Documents/"
sending incremental file list
binary/
binary/Eula.txt
binary/cherrytree.lnk
binary/depends64.chm
binary/depends64.dll
binary/depends64.exe
binary/mtputty.exe
binary/procexp.chm
binary/procexp.exe
binary/pscp.exe
binary/putty.exe
binary/sqlite3.exe
binary/wget.exe
sent 4115294 bytes received 244 bytes 1175868.00 bytes/sec
total size is 8036311 speedup is 1.95
c:\Rtools\bin>rm c:\users\limingc\Documents\binary\procexp.exe
cygwin warning:
MS-DOS style path detected: c:\users\limingc\Documents\binary\procexp.exe
Preferred POSIX equivalent is: /cygdrive/c/users/limingc/Documents/binary/procexp.exe
CYGWIN environment variable option "nodosfilewarning" turns off this warning.
Consult the user's guide for more details about POSIX paths:
http://cygwin.com/cygwin-ug-net/using.html#using-pathnames
c:\Rtools\bin>rsync -avz "/cygdrive/c/users/limingc/Downloads/binary" "/cygdrive/c/users/limingc/Documents/"
sending incremental file list
binary/
binary/procexp.exe
sent 1767277 bytes received 35 bytes 3534624.00 bytes/sec
total size is 8036311 speedup is 4.55
c:\Rtools\bin>
Unforunately, if the destination is a network drive, I could get a permission denied (13) error. See also http://superuser.com/questions/69620/rsync-file-permissions-on-windows
Install rgdal package (geospatial Data) on ubuntu
Terminal
sudo apt-get install libgdal1-dev libproj-dev
R
install.packages("rgdal")
Set up Emacs on Windows
Edit the file C:\Program Files\GNU Emacs 23.2\site-lisp\site-start.el with something like
(setq-default inferior-R-program-name
"c:/program files/r/r-2.15.2/bin/i386/rterm.exe")
Database
RSQLite
- https://cran.r-project.org/web/packages/RSQLite/vignettes/RSQLite.html
- https://github.com/rstats-db/RSQLite
Creating a new database:
library(DBI)
mydb <- dbConnect(RSQLite::SQLite(), "my-db.sqlite")
dbDisconnect(mydb)
unlink("my-db.sqlite")
# temporary database
mydb <- dbConnect(RSQLite::SQLite(), "")
dbDisconnect(mydb)
Loading data:
mydb <- dbConnect(RSQLite::SQLite(), "") dbWriteTable(mydb, "mtcars", mtcars) dbWriteTable(mydb, "iris", iris) dbListTables(mydb) dbListFields(con, "mtcars") dbReadTable(con, "mtcars")
Queries:
dbGetQuery(mydb, 'SELECT * FROM mtcars LIMIT 5') dbGetQuery(mydb, 'SELECT * FROM iris WHERE "Sepal.Length" < 4.6') dbGetQuery(mydb, 'SELECT * FROM iris WHERE "Sepal.Length" < :x', params = list(x = 4.6)) res <- dbSendQuery(con, "SELECT * FROM mtcars WHERE cyl = 4") dbFetch(res)
Batched queries:
dbClearResult(rs)
rs <- dbSendQuery(mydb, 'SELECT * FROM mtcars')
while (!dbHasCompleted(rs)) {
df <- dbFetch(rs, n = 10)
print(nrow(df))
}
dbClearResult(rs)
Multiple parameterised queries:
rs <- dbSendQuery(mydb, 'SELECT * FROM iris WHERE "Sepal.Length" = :x') dbBind(rs, param = list(x = seq(4, 4.4, by = 0.1))) nrow(dbFetch(rs)) #> [1] 4 dbClearResult(rs)
Statements:
dbExecute(mydb, 'DELETE FROM iris WHERE "Sepal.Length" < 4') #> [1] 0 rs <- dbSendStatement(mydb, 'DELETE FROM iris WHERE "Sepal.Length" < :x') dbBind(rs, param = list(x = 4.5)) dbGetRowsAffected(rs) #> [1] 4 dbClearResult(rs)
sqldf
Manipulate R data frames using SQL. Depends on RSQLite. A use of gsub, reshape2 and sqldf with healthcare data
RPostgreSQL
RMySQL
- http://datascienceplus.com/bringing-the-powers-of-sql-into-r/
- See here about the installation of the required package (libmysqlclient-dev) in Ubuntu.
MongoDB
- http://www.r-bloggers.com/r-and-mongodb/
- http://watson.nci.nih.gov/~sdavis/blog/rmongodb-using-R-with-mongo/
odbc
RODBC
DBI
dbplyr
- To use databases with dplyr, you need to first install dbplyr
- https://db.rstudio.com/dplyr/
- Five commonly used backends: RMySQL, RPostgreSQ, RSQLite, ODBC, bigrquery.
- http://www.datacarpentry.org/R-ecology-lesson/05-r-and-databases.html
Create a new SQLite database:
surveys <- read.csv("data/surveys.csv")
plots <- read.csv("data/plots.csv")
my_db_file <- "portal-database.sqlite"
my_db <- src_sqlite(my_db_file, create = TRUE)
copy_to(my_db, surveys)
copy_to(my_db, plots)
my_db
Connect to a database:
download.file(url = "https://ndownloader.figshare.com/files/2292171",
destfile = "portal_mammals.sqlite", mode = "wb")
library(dbplyr)
library(dplyr)
mammals <- src_sqlite("portal_mammals.sqlite")
Querying the database with the SQL syntax:
tbl(mammals, sql("SELECT year, species_id, plot_id FROM surveys"))
Querying the database with the dplyr syntax:
surveys <- tbl(mammals, "surveys")
surveys %>%
select(year, species_id, plot_id)
head(surveys, n = 10)
show_query(head(surveys, n = 10)) # show which SQL commands are actually sent to the database
Simple database queries:
surveys %>% filter(weight < 5) %>% select(species_id, sex, weight)
Laziness (instruct R to stop being lazy):
data_subset <- surveys %>% filter(weight < 5) %>% select(species_id, sex, weight) %>% collect()
Complex database queries:
plots <- tbl(mammals, "plots") plots # # The plot_id column features in the plots table surveys # The plot_id column also features in the surveys table # Join databases method 1 plots %>% filter(plot_id == 1) %>% inner_join(surveys) %>% collect()
NoSQL
nodbi: the NoSQL Database Connector
Github
R source
https://github.com/wch/r-source/ Daily update, interesting, should be visited every day. Clicking 1000+ commits to look at daily changes.
If we are interested in a certain branch (say 3.2), look for R-3-2-branch.
R packages (only) source (metacran)
- https://github.com/cran/ by Gábor Csárdi, the author of igraph software.
Bioconductor packages source
Announcement, https://github.com/Bioconductor-mirror
Send local repository to Github in R by using reports package
http://www.youtube.com/watch?v=WdOI_-aZV0Y
My collection
- https://github.com/arraytools
- https://gist.github.com/4383351 heatmap using leukemia data
- https://gist.github.com/4382774 heatmap using sequential data
- https://gist.github.com/4484270 biocLite
How to download
Clone ~ Download.
- Command line
git clone https://gist.github.com/4484270.git
This will create a subdirectory called '4484270' with all cloned files there.
- Within R
library(devtools)
source_gist("4484270")
or First download the json file from
https://api.github.com/users/MYUSERLOGIN/gists
and then
library(RJSONIO)
x <- fromJSON("~/Downloads/gists.json")
setwd("~/Downloads/")
gist.id <- lapply(x, "[[", "id")
lapply(gist.id, function(x){
cmd <- paste0("git clone https://gist.github.com/", x, ".git")
system(cmd)
})
Jekyll
An Easy Start with Jekyll, for R-Bloggers
Connect R with Arduino
- http://lamages.blogspot.com/2012/10/connecting-real-world-to-r-with-arduino.html
- http://jean-robert.github.io/2012/11/11/thermometer-R-using-Arduino-Java.html
- http://bio7.org/?p=2049
- http://www.rforge.net/Arduino/svn.html
Android App
- R Instructor $4.84
- Statistical Distribution (Not R related app)
Common plots tips
Grouped boxplots
- Box Plots of Two-Way Layout
- Step by step to create a grouped boxplots
- 'at' parameter in boxplot() to change the equal spaced boxplots
- embed par(mar=) in boxplot()
- mtext(line=) to solve the problem the xlab overlapped with labels.
- ggplot2 approach (Hint: facet_grid is used)
Weather Time Line
The plot looks similar to a boxplot though it is not. See a screenshot on Android by Sam Ruston.
Horizontal bar plot
library(ggplot2)
dtf <- data.frame(x = c("ETB", "PMA", "PER", "KON", "TRA",
"DDR", "BUM", "MAT", "HED", "EXP"),
y = c(.02, .11, -.01, -.03, -.03, .02, .1, -.01, -.02, 0.06))
ggplot(dtf, aes(x, y)) +
geom_bar(stat = "identity", aes(fill = x), show.legend = FALSE) +
coord_flip() + xlab("") + ylab("Fold Change")
Include bar values in a barplot
- https://stats.stackexchange.com/questions/3879/how-to-put-values-over-bars-in-barplot-in-r.
- barplot(), text() and axis() functions. The data can be from a table() object.
- How to label a barplot bar with positive and negative bars with ggplot2
Use text().
Or use geom_text() if we are using the ggplot2 package. See an example here or this.
For stacked barplot, see this post.
Grouped barplots
- https://www.r-graph-gallery.com/barplot/, https://www.r-graph-gallery.com/48-grouped-barplot-with-ggplot2/ (simpliest, no error bars)
library(ggplot2) # mydata <- data.frame(OUTGRP, INGRP, value) ggplot(mydata, aes(fill=INGRP, y=value, x=OUTGRP)) + geom_bar(position="dodge", stat="identity") - https://datascienceplus.com/building-barplots-with-error-bars/. The error bars define 2 se (95% interval) for the black-and-white version and 1 se (68% interval) for ggplots. Be careful.
> 1 - 2*(1-pnorm(1)) [1] 0.6826895 > 1 - 2*(1-pnorm(1.96)) [1] 0.9500042
- two bars in one factor (stack). The data can be a 2-dim matrix with numerical values.
- two bars in one factor, Drawing multiple barplots on a graph in R (next to each other)
- Three variables barplots
- More alternatives (not done by R)
Math expression
- ?plotmath
- https://stackoverflow.com/questions/4973898/combining-paste-and-expression-functions-in-plot-labels
- http://vis.supstat.com/2013/04/mathematical-annotation-in-r/
- https://andyphilips.github.io/blog/2017/08/16/mathematical-symbols-in-r-plots.html
# Expressions
plot(x,y, xlab = expression(hat(x)[t]),
ylab = expression(phi^{rho + a}),
main = "Pure Expressions")
# Expressions with Spacing
# '~' is to add space and '*' is to squish characters together
plot(1:10, xlab= expression(Delta * 'C'))
plot(x,y, xlab = expression(hat(x)[t] ~ z ~ w),
ylab = expression(phi^{rho + a} * z * w),
main = "Pure Expressions with Spacing")
# Expressions with Text
plot(x,y,
xlab = expression(paste("Text here ", hat(x), " here ", z^rho, " and here")),
ylab = expression(paste("Here is some text of ", phi^{rho})),
main = "Expressions with Text")
# Substituting Expressions
plot(x,y,
xlab = substitute(paste("Here is ", pi, " = ", p), list(p = py)),
ylab = substitute(paste("e is = ", e ), list(e = ee)),
main = "Substituted Expressions")
Rotating x axis labels for barplot
https://stackoverflow.com/questions/10286473/rotating-x-axis-labels-in-r-for-barplot
barplot(mytable,main="Car makes",ylab="Freqency",xlab="make",las=2)
Set R plots x axis to show at y=0
https://stackoverflow.com/questions/3422203/set-r-plots-x-axis-to-show-at-y-0
plot(1:10, rnorm(10), ylim=c(0,10), yaxs="i")
Different colors of axis labels in barplot
See Vary colors of axis labels in R based on another variable
Method 1: Append labels for the 2nd, 3rd, ... color gradually because 'col.axis' argument cannot accept more than one color.
tN <- table(Ni <- stats::rpois(100, lambda = 5)) r <- barplot(tN, col = rainbow(20)) axis(1, 1, LETTERS[1], col.axis="red", col="red") axis(1, 2, LETTERS[2], col.axis="blue", col = "blue")
Method 2: text() which can accept multiple colors in 'col' parameter but we need to find out the (x, y) by ourselves.
barplot(tN, col = rainbow(20), axisnames = F)
text(4:6, par("usr")[3]-2 , LETTERS[4:6], col=c("black","red","blue"), xpd=TRUE)
Use text() to draw labels on X/Y-axis including rotation
- adj = 1 means top/rigth alignment. The default is to center the text.
- par("usr") gives the extremes of the user coordinates of the plotting region of the form c(x1, x2, y1, y2).
- par("usr") is determined *after* a plot has been created
- Example of using the "usr" parameter
- https://datascienceplus.com/building-barplots-with-error-bars/
par(mar = c(5, 6, 4, 5) + 0.1)
plot(..., xaxt = "n") # "n" suppresses plotting of the axis; need mtext() and axis() to supplement
text(x = barCenters, y = par("usr")[3] - 1, srt = 45,
adj = 1, labels = myData$names, xpd = TRUE)
Vertically stacked plots with the same x axis
Superimpose a density plot or any curves
Use lines().
Example 1
plot(cars, main = "Stopping Distance versus Speed") lines(stats::lowess(cars))
Example 2
require(survival)
n = 10000
beta1 = 2; beta2 = -1
lambdaT = 1 # baseline hazard
lambdaC = 2 # hazard of censoring
set.seed(1234)
x1 = rnorm(n,0)
x2 = rnorm(n,0)
# true event time
T = rweibull(n, shape=1, scale=lambdaT*exp(-beta1*x1-beta2*x2))
C <- rweibull(n, shape=1, scale=lambdaC)
time = pmin(T,C)
status <- 1*(T <= C)
status2 <- 1-status
plot(survfit(Surv(time, status2) ~ 1),
ylab="Survival probability",
main = 'Exponential censoring time')
xseq <- seq(.1, max(time), length =100)
func <- function(x) 1-pweibull(x, shape = 1, scale = lambdaC)
lines(xseq, func(xseq), col = 'red') # survival function of Weibull
Time series
Time series stock price plot
- http://blog.revolutionanalytics.com/2015/08/plotting-time-series-in-r.html (ggplot2, xts, dygraphs)
- Visualize your Portfolio’s Performance and Generate a Nice Report with R
- https://timelyportfolio.github.io/rCharts_time_series/history.html
library(quantmod)
getSymbols("AAPL")
getSymbols("IBM") # similar to AAPL
getSymbols("CSCO") # much smaller than AAPL, IBM
getSymbols("DJI") # Dow Jones, huge
chart_Series(Cl(AAPL), TA="add_TA(Cl(IBM), col='blue', on=1); add_TA(Cl(CSCO), col = 'green', on=1)",
col='orange', subset = '2017::2017-08')
tail(Cl(DJI))
Timeline plot
Circular plot
- http://freakonometrics.hypotheses.org/20667 which uses https://cran.r-project.org/web/packages/circlize/ circlize] package.
- https://www.biostars.org/p/17728/
- RCircos package from CRAN.
- OmicCircos from Bioconductor.
Word cloud
- Text mining and word cloud fundamentals in R : 5 simple steps you should know
- 7 Alternatives to Word Clouds for Visualizing Long Lists of Data
- Data + Art STEAM Project: Initial Results
World map
Visualising SSH attacks with R (rworldmap and rgeolocate packages)
Diagram/flowchart
DiagrammeR
http://rich-iannone.github.io/DiagrammeR/
diagram
Functions for Visualising Simple Graphs (Networks), Plotting Flow Diagrams
Venn Diagram
- limma http://www.ats.ucla.edu/stat/r/faq/venn.htm - only black and white?
- VennDiagram - input has to be the numbers instead of the original vector?
- http://manuals.bioinformatics.ucr.edu/home/R_BioCondManual#TOC-Venn-Diagrams and the R code or the Bioc package systemPipeR
# systemPipeR package method
library(systemPipeR)
setlist <- list(A=sample(letters, 18), B=sample(letters, 16), C=sample(letters, 20), D=sample(letters, 22), E=sample(letters, 18))
OLlist <- overLapper(setlist[1:3], type="vennsets")
vennPlot(list(OLlist))
# R script source method
source("http://faculty.ucr.edu/~tgirke/Documents/R_BioCond/My_R_Scripts/overLapper.R")
setlist <- list(A=sample(letters, 18), B=sample(letters, 16), C=sample(letters, 20), D=sample(letters, 22), E=sample(letters, 18))
# or (obtained by dput(setlist))
setlist <- structure(list(A = c("o", "h", "u", "p", "i", "s", "a", "w",
"b", "z", "n", "c", "k", "j", "y", "m", "t", "q"), B = c("h",
"r", "x", "y", "b", "t", "d", "o", "m", "q", "g", "v", "c", "u",
"f", "z"), C = c("b", "e", "t", "u", "s", "j", "o", "k", "d",
"l", "g", "i", "w", "n", "p", "a", "y", "x", "m", "z"), D = c("f",
"g", "b", "k", "j", "m", "e", "q", "i", "d", "o", "l", "c", "t",
"x", "r", "s", "u", "w", "a", "z", "n"), E = c("u", "w", "o",
"k", "n", "h", "p", "z", "l", "m", "r", "d", "q", "s", "x", "b",
"v", "t"), F = c("o", "j", "r", "c", "l", "l", "u", "b", "f",
"d", "u", "m", "y", "t", "y", "s", "a", "g", "t", "m", "x", "m"
)), .Names = c("A", "B", "C", "D", "E", "F"))
OLlist <- overLapper(setlist[1:3], type="vennsets")
counts <- list(sapply(OLlist$Venn_List, length))
vennPlot(counts=counts)
Bump chart/Metro map
https://dominikkoch.github.io/Bump-Chart/
Amazing plots
New R logo 2/11/2016
- http://rud.is/b/2016/02/11/plot-the-new-svg-r-logo-with-ggplot2/
- https://www.stat.auckland.ac.nz/~paul/Reports/Rlogo/Rlogo.html
library(sp)
library(maptools)
library(ggplot2)
library(ggthemes)
# rgeos requires the installation of GEOS from http://trac.osgeo.org/geos/
system("curl http://download.osgeo.org/geos/geos-3.5.0.tar.bz2 | tar jx")
system("cd geos-3.5.0; ./configure; make; sudo make install")
library(rgeos)
r_wkt_gist_file <- "https://gist.githubusercontent.com/hrbrmstr/07d0ccf14c2ff109f55a/raw/db274a39b8f024468f8550d7aeaabb83c576f7ef/rlogo.wkt"
if (!file.exists("rlogo.wkt")) download.file(r_wkt_gist_file, "rlogo.wkt")
rlogo <- readWKT(paste0(readLines("rlogo.wkt", warn=FALSE))) # rgeos
rlogo_shp <- SpatialPolygonsDataFrame(rlogo, data.frame(poly=c("halo", "r"))) # sp
rlogo_poly <- fortify(rlogo_shp, region="poly") # ggplot2
ggplot(rlogo_poly) +
geom_polygon(aes(x=long, y=lat, group=id, fill=id)) +
scale_fill_manual(values=c(halo="#b8babf", r="#1e63b5")) +
coord_equal() +
theme_map() +
theme(legend.position="none")
3D plot
Using persp function to create the following plot.
### Random pattern
# Create matrix with random values with dimension of final grid
rand <- rnorm(441, mean=0.3, sd=0.1)
mat.rand <- matrix(rand, nrow=21)
# Create another matrix for the colors. Start by making all cells green
fill <- matrix("green3", nr = 21, nc = 21)
# Change colors in each cell based on corresponding mat.rand value
fcol <- fill
fcol[] <- terrain.colors(40)[cut(mat.rand,
stats::quantile(mat.rand, seq(0,1, len = 41),
na.rm=T), include.lowest = TRUE)]
# Create concave surface using expontential function
x <- -10:10
y <- x^2
y <- as.matrix(y)
y1 <- y
for(i in 1:20){tmp <- cbind(y,y1); y1 <- tmp[,1]; y <- tmp;}
mat <- tmp[1:21, 1:21]
# Plot it up!
persp(1:21, 1:21, t(mat)/10, theta = 90, phi = 35,col=fcol,
scale = FALSE, axes = FALSE, box = FALSE)
### Organized pattern
# Same as before
rand <- rnorm(441, mean=0.3, sd=0.1)
# Create concave surface using expontential function
x <- -10:10
y <- x^2
y <- as.matrix(y)
for(i in 1:20){tmp <- cbind(y,y); y1 <- tmp[,1]; y <- tmp;}
mat <- tmp[1:21, 1:21]
###Organize rand by y and put into matrix form
o <- order(rand,as.vector(mat))
o.tmp <- cbind(rand[o], rev(sort(as.vector(mat))))
mat.org <- matrix(o.tmp[,1], nrow=21)
half.1 <- mat.org[,seq(1,21,2)]
half.2 <- mat.org[,rev(seq(2,20,2))]
full <- cbind(half.1, half.2)
full <- t(full)
# Again, create color matrix and populate using rand values
zi <- full[-1, -1] + full[-1, -21] + full[-21,-1] + full[-21, -21]
fill <- matrix("green3", nr = 20, nc = 20)
fcol <- fill
fcol[] <- terrain.colors(40)[cut(zi,
stats::quantile(zi, seq(0,1, len = 41), na.rm=T),
include.lowest = TRUE)]
# Plot it up!
persp(1:21, 1:21, t(mat)/10, theta = 90, phi = 35,col=t(fcol),
scale = FALSE, axes = FALSE, box = FALSE)
Christmas tree
http://wiekvoet.blogspot.com/2014/12/merry-christmas.html
# http://blogs.sas.com/content/iml/2012/12/14/a-fractal-christmas-tree/
# Each row is a 2x2 linear transformation
# Christmas tree
L <- matrix(
c(0.03, 0, 0 , 0.1,
0.85, 0.00, 0.00, 0.85,
0.8, 0.00, 0.00, 0.8,
0.2, -0.08, 0.15, 0.22,
-0.2, 0.08, 0.15, 0.22,
0.25, -0.1, 0.12, 0.25,
-0.2, 0.1, 0.12, 0.2),
nrow=4)
# ... and each row is a translation vector
B <- matrix(
c(0, 0,
0, 1.5,
0, 1.5,
0, 0.85,
0, 0.85,
0, 0.3,
0, 0.4),
nrow=2)
prob = c(0.02, 0.6,.08, 0.07, 0.07, 0.07, 0.07)
# Iterate the discrete stochastic map
N = 1e5 #5 # number of iterations
x = matrix(NA,nrow=2,ncol=N)
x[,1] = c(0,2) # initial point
k <- sample(1:7,N,prob,replace=TRUE) # values 1-7
for (i in 2:N)
x[,i] = crossprod(matrix(L[,k[i]],nrow=2),x[,i-1]) + B[,k[i]] # iterate
# Plot the iteration history
png('card.png')
par(bg='darkblue',mar=rep(0,4))
plot(x=x[1,],y=x[2,],
col=grep('green',colors(),value=TRUE),
axes=FALSE,
cex=.1,
xlab='',
ylab='' )#,pch='.')
bals <- sample(N,20)
points(x=x[1,bals],y=x[2,bals]-.1,
col=c('red','blue','yellow','orange'),
cex=2,
pch=19
)
text(x=-.7,y=8,
labels='Merry',
adj=c(.5,.5),
srt=45,
vfont=c('script','plain'),
cex=3,
col='gold'
)
text(x=0.7,y=8,
labels='Christmas',
adj=c(.5,.5),
srt=-45,
vfont=c('script','plain'),
cex=3,
col='gold'
)
Happy Thanksgiving
Happy Valentine's Day
https://rud.is/b/2017/02/14/geom%E2%9D%A4%EF%B8%8F/
treemap
Voronoi diagram
- https://www.stat.auckland.ac.nz/~paul/Reports/VoronoiTreemap/voronoiTreeMap.html
- http://letstalkdata.com/2014/05/creating-voronoi-diagrams-with-ggplot/
Silent Night
# https://aschinchon.wordpress.com/2014/03/13/the-lonely-acacia-is-rocked-by-the-wind-of-the-african-night/
depth <- 9
angle<-30 #Between branches division
L <- 0.90 #Decreasing rate of branches by depth
nstars <- 300 #Number of stars to draw
mstars <- matrix(runif(2*nstars), ncol=2)
branches <- rbind(c(1,0,0,abs(jitter(0)),1,jitter(5, amount = 5)), data.frame())
colnames(branches) <- c("depth", "x1", "y1", "x2", "y2", "inertia")
for(i in 1:depth)
{
df <- branches[branches$depth==i,]
for(j in 1:nrow(df))
{
branches <- rbind(branches, c(df[j,1]+1, df[j,4], df[j,5], df[j,4]+L^(2*i+1)*sin(pi*(df[j,6]+angle)/180),
df[j,5]+L^(2*i+1)*cos(pi*(df[j,6]+angle)/180), df[j,6]+angle+jitter(10, amount = 8)))
branches <- rbind(branches, c(df[j,1]+1, df[j,4], df[j,5], df[j,4]+L^(2*i+1)*sin(pi*(df[j,6]-angle)/180),
df[j,5]+L^(2*i+1)*cos(pi*(df[j,6]-angle)/180), df[j,6]-angle+jitter(10, amount = 8)))
}
}
nodes <- rbind(as.matrix(branches[,2:3]), as.matrix(branches[,4:5]))
png("image.png", width = 1200, height = 600)
plot.new()
par(mai = rep(0, 4), bg = "gray12")
plot(nodes, type="n", xlim=c(-7, 3), ylim=c(0, 5))
for (i in 1:nrow(mstars))
{
points(x=10*mstars[i,1]-7, y=5*mstars[i,2], col = "blue4", cex=.7, pch=16)
points(x=10*mstars[i,1]-7, y=5*mstars[i,2], col = "blue", cex=.3, pch=16)
points(x=10*mstars[i,1]-7, y=5*mstars[i,2], col = "white", cex=.1, pch=16)
}
# The moon
points(x=-5, y=3.5, cex=40, pch=16, col="lightyellow")
# The tree
for (i in 1:nrow(branches)) {
lines(x=branches[i,c(2,4)], y=branches[i,c(3,5)],
col = paste("gray", as.character(sample(seq(from=50, to=round(50+5*branches[i,1]), by=1), 1)), sep = ""),
lwd=(65/(1+3*branches[i,1])))
}
rm(branches)
dev.off()
The Travelling Salesman Portrait
https://fronkonstin.com/2018/04/04/the-travelling-salesman-portrait/
Moon phase calendar
https://chichacha.netlify.com/2018/05/26/making-calendar-with-ggplot-moon-phase-calendar/
Google Analytics
GAR package
http://www.analyticsforfun.com/2015/10/query-your-google-analytics-data-with.html
Linear Programming
http://www.r-bloggers.com/modeling-and-solving-linear-programming-with-r-free-book/
Amazon Alexa
R and Singularity
https://www.rstudio.com/rviews/2017/03/29/r-and-singularity/
Teach kids about R with Minecraft
http://blog.revolutionanalytics.com/2017/06/teach-kids-about-r-with-minecraft.html
Secure API keys
Securely store API keys in R scripts with the "secret" package
Vision and image recognition
- https://www.stoltzmaniac.com/google-vision-api-in-r-rooglevision/ Google vision API IN R] – RoogleVision
- Computer Vision Algorithms for R users and https://github.com/bnosac/image
Turn pictures into coloring pages
https://gist.github.com/jeroen/53a5f721cf81de2acba82ea47d0b19d0
Numerical optimization
- uniroot: One Dimensional Root (Zero) Finding. This is used in simulating survival data for predefined censoring rate
- optimize: One Dimensional Optimization
- optim: General-purpose optimization based on Nelder–Mead, quasi-Newton and conjugate-gradient algorithms.
- constrOptim: Linearly Constrained Optimization
- nlm: Non-Linear Minimization
- nls: Nonlinear Least Squares
R packages
R package management
Packages loaded at startup
getOption("defaultPackages")
- available.packages(); see packageStatus().
- download.packages()
- packageStatus(), update(), upgrade(). packageStatus() will return a list with two components:
- inst - a data frame with columns as the matrix returned by installed.packages plus "Status", a factor with levels c("ok", "upgrade"). Note: the manual does not mention "unavailable" case (but I do get it) in R 3.2.0?
- avail - a data frame with columns as the matrix returned by available.packages plus "Status", a factor with levels c("installed", "not installed", "unavailable"). Note: I don't get the "unavailable" case in R 3.2.0?
> x <- packageStatus()
> names(x)
[1] "inst" "avail"
> dim(x[['inst']])
[1] 225 17
> x[['inst']][1:3, ]
Package LibPath Version Priority Depends Imports
acepack acepack C:/Program Files/R/R-3.1.2/library 1.3-3.3 <NA> <NA> <NA>
adabag adabag C:/Program Files/R/R-3.1.2/library 4.0 <NA> rpart, mlbench, caret <NA>
affxparser affxparser C:/Program Files/R/R-3.1.2/library 1.38.0 <NA> R (>= 2.6.0) <NA>
LinkingTo Suggests Enhances
acepack <NA> <NA> <NA>
adabag <NA> <NA> <NA>
affxparser <NA> R.oo (>= 1.18.0), R.utils (>= 1.32.4),\nAffymetrixDataTestFiles <NA>
License License_is_FOSS License_restricts_use OS_type MD5sum NeedsCompilation Built
acepack MIT + file LICENSE <NA> <NA> <NA> <NA> yes 3.1.2
adabag GPL (>= 2) <NA> <NA> <NA> <NA> no 3.1.2
affxparser LGPL (>= 2) <NA> <NA> <NA> <NA> <NA> 3.1.1
Status
acepack ok
adabag ok
affxparser unavailable
> dim(x[['avail']])
[1] 6538 18
> x[['avail']][1:3, ]
Package Version Priority Depends Imports LinkingTo
A3 A3 0.9.2 <NA> R (>= 2.15.0), xtable, pbapply <NA> <NA>
ABCExtremes ABCExtremes 1.0 <NA> SpatialExtremes, combinat <NA> <NA>
ABCanalysis ABCanalysis 1.0.1 <NA> R (>= 2.10) Hmisc, plotrix <NA>
Suggests Enhances License License_is_FOSS License_restricts_use OS_type Archs
A3 randomForest, e1071 <NA> GPL (>= 2) <NA> <NA> <NA> <NA>
ABCExtremes <NA> <NA> GPL-2 <NA> <NA> <NA> <NA>
ABCanalysis <NA> <NA> GPL-3 <NA> <NA> <NA> <NA>
MD5sum NeedsCompilation File Repository Status
A3 <NA> <NA> <NA> http://cran.rstudio.com/bin/windows/contrib/3.1 not installed
ABCExtremes <NA> <NA> <NA> http://cran.rstudio.com/bin/windows/contrib/3.1 not installed
ABCanalysis <NA> <NA> <NA> http://cran.rstudio.com/bin/windows/contrib/3.1 not installed
- packageVersion(), packageDescription()
- install.packages(), remove.packages().
- installed.packages(); see packageStatus().
- update.packages(), old.packages(), new.packages()
- setRepositories()
- contrib.url()
- chooseCRANmirror(), chooseBioCmirror()
- suppressForeignCheck()
install.packages()
By default, install.packages() will check versions and install uninstalled packages shown in 'Depends', 'Imports', and 'LinkingTo' fields. See R-exts manual.
If we want to install packages listed in 'Suggests' field, we should specify it explicitly by using dependencies argument:
install.packages(XXXX, dependencies = c("Depends", "Imports", "Suggests", "LinkingTo"))
# OR
install.packages(XXXX, dependencies = TRUE)
For example, if I use a plain install.packages() command to install downloader package
install.packages("downloader")
it will only install 'digest' and 'downloader' packages. If I use
install.packages("downloader", dependencies=TRUE)
it will also install 'testhat' package.
The install.packages function source code can be found in R -> src -> library -> utils -> R -> packages2.R file from Github repository (put 'install.packages' in the search box).
Install and load the package at the same time
p_load() function from the pacman package.
An example of installing the purrr package.
Check installed Bioconductor version
Following this post, use tools:::.BioC_version_associated_with_R_version().
Mind the '.' in front of the 'BioC'. It may be possible for some installed packages to have been sourced from a different BioC version.
tools:::.BioC_version_associated_with_R_version() # `3.6' tools:::.BioC_version_associated_with_R_version() == '3.6' # TRUE
CRAN Package Depends on Bioconductor Package
For example, if I run install.packages("NanoStringNorm") to install the package from CRAN, I may get
ERROR: dependency ‘vsn’ is not available for package ‘NanoStringNorm’
This is because the NanoStringNorm package depends on the vsn package which is on Bioconductor.
Another instance is CRAN's 'biospear' depends on Bioc's 'survcomp'.
One solution is to run a line setRepositories(ind=1:2). See this post or this one. Note that the default repository list can be found at (Ubuntu) /usr/lib/R/etc/repositories file.
options("repos") # display the available repositories (only CRAN)
setRepositories(ind=1:2)
options("repos") # CRAN and bioc are included
# CRAN
# "https://cloud.r-project.org"
# "https://bioconductor.org/packages/3.6/bioc"
install.packages("biospear") # it will prompt to select CRAN
install.packages("biospear", repos = "http://cran.rstudio.com") # NOT work since bioc repos is erased
This will also install the BiocInstaller package if it has not been installed before. See also Install Bioconductor Packages.
install a tar.gz (e.g. an archived package) from a local directory
R CMD INSTALL <package-name>.tar.gz
Or in R:
install(<pathtopackage>) # this will use 'R CMD INSTALL' to install the package.
# It will try to install dependencies of the package from CRAN,
# if they're not already installed.
install.packages(<pathtopackage>, repos = NULL)
The installation process can be nasty due to the dependency issue. Consider the 'biospear' package
biospear - plsRcox (archived) - plsRglm (archived) - bipartite
- lars
- pls
- kernlab
- mixOmics
- risksetROC
- survcomp (Bioconductor)
- rms
So in order to install the 'plsRcox' package, we need to do the following steps. Note: plsRcox package is back on 6/2/2018.
# For curl
system("apt update")
system("apt install curl libcurl4-openssl-dev libssl-dev")
# For X11
system("apt install libcgal-dev libglu1-mesa-dev libglu1-mesa-dev")
system("apt install libfreetype6-dev") # https://stackoverflow.com/questions/31820865/error-in-installing-rgl-package
source("https://bioconductor.org/biocLite.R")
biocLite("survcomp") # this has to be run before the next command of installing a bunch of packages from CRAN
install.packages("https://cran.r-project.org/src/contrib/Archive/biospear/biospear_1.0.1.tar.gz",
repos = NULL, type="source")
# ERROR: dependencies ‘pkgconfig’, ‘cobs’, ‘corpcor’, ‘devtools’, ‘glmnet’, ‘grplasso’, ‘mboost’, ‘plsRcox’,
# ‘pROC’, ‘PRROC’, ‘RCurl’, ‘survAUC’ are not available for package ‘biospear’
install.packages(c("pkgconfig", "cobs", "corpcor", "devtools", "glmnet", "grplasso", "mboost",
"plsRcox", "pROC", "PRROC", "RCurl", "survAUC"))
# optional: install.packages(c("doRNG", "mvnfast"))
install.packages("https://cran.r-project.org/src/contrib/Archive/biospear/biospear_1.0.1.tar.gz",
repos = NULL, type="source")
# OR
# devtools::install_github("cran/biospear")
library(biospear) # verify
To install the (deprecated, bioc) packages 'inSilicoMerging',
biocLite(c('rjson', 'Biobase', 'RCurl'))
# destination directory is required
# download.file("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoDb_2.7.0.tar.gz",
# "~/Downloads/inSilicoDb_2.7.0.tar.gz")
# download.file("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoMerging_1.15.0.tar.gz",
# "~/Downloads/inSilicoMerging_1.15.0.tar.gz")
# ~/Downloads or $HOME/Downloads won't work in untar()
# untar("~/Downloads/inSilicoDb_2.7.0.tar.gz", exdir="/home/brb/Downloads")
# untar("~/Downloads/inSilicoMerging_1.15.0.tar.gz", exdir="/home/brb/Downloads")
# install.packages("~/Downloads/inSilicoDb", repos = NULL)
# install.packages("~/Downloads/inSilicoMerging", repos = NULL)
install.packages("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoDb_2.7.0.tar.gz",
repos = NULL, type = "source")
install.packages("http://www.bioconductor.org/packages/3.3/bioc/src/contrib/inSilicoMerging_1.15.0.tar.gz",
repos = NULL, type = "source")
Query an R package installed locally
packageDescription("MASS")
packageVersion("MASS")
Query an R package (from CRAN) basic information
packageStatus() # Summarize information about installed packages available.packages() # List Available Packages at CRAN-like Repositories
The available.packages() command is useful for understanding package dependency. Use setRepositories() or 'RGUI -> Packages -> select repositories' to select repositories and options()$repos to check or change the repositories.
Also the packageStatus() is another useful function for query how many packages are in the repositories, how many have been installed, and individual package status (installed or not, needs to be upgraded or not).
> options()$repos
CRAN
"https://cran.rstudio.com/"
> packageStatus()
Number of installed packages:
ok upgrade unavailable
C:/Program Files/R/R-3.0.1/library 110 0 1
Number of available packages (each package counted only once):
installed not installed
http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0 76 4563
http://www.stats.ox.ac.uk/pub/RWin/bin/windows/contrib/3.0 0 5
http://www.bioconductor.org/packages/2.12/bioc/bin/windows/contrib/3.0 16 625
http://www.bioconductor.org/packages/2.12/data/annotation/bin/windows/contrib/3.0 4 686
> tmp <- available.packages()
> str(tmp)
chr [1:5975, 1:17] "A3" "ABCExtremes" "ABCp2" "ACCLMA" "ACD" "ACNE" "ADGofTest" "ADM3" "AER" ...
- attr(*, "dimnames")=List of 2
..$ : chr [1:5975] "A3" "ABCExtremes" "ABCp2" "ACCLMA" ...
..$ : chr [1:17] "Package" "Version" "Priority" "Depends" ...
> tmp[1:3,]
Package Version Priority Depends Imports LinkingTo Suggests
A3 "A3" "0.9.2" NA "xtable, pbapply" NA NA "randomForest, e1071"
ABCExtremes "ABCExtremes" "1.0" NA "SpatialExtremes, combinat" NA NA NA
ABCp2 "ABCp2" "1.1" NA "MASS" NA NA NA
Enhances License License_is_FOSS License_restricts_use OS_type Archs MD5sum NeedsCompilation File
A3 NA "GPL (>= 2)" NA NA NA NA NA NA NA
ABCExtremes NA "GPL-2" NA NA NA NA NA NA NA
ABCp2 NA "GPL-2" NA NA NA NA NA NA NA
Repository
A3 "http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0"
ABCExtremes "http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0"
ABCp2 "http://watson.nci.nih.gov/cran_mirror/bin/windows/contrib/3.0"
And the following commands find which package depends on Rcpp and also which are from bioconductor repository.
> pkgName <- "Rcpp"
> rownames(tmp)[grep(pkgName, tmp[,"Depends"])]
> tmp[grep("Rcpp", tmp[,"Depends"]), "Depends"]
> ind <- intersect(grep(pkgName, tmp[,"Depends"]), grep("bioconductor", tmp[, "Repository"]))
> rownames(grep)[ind]
NULL
> rownames(tmp)[ind]
[1] "ddgraph" "DESeq2" "GeneNetworkBuilder" "GOSemSim" "GRENITS"
[6] "mosaics" "mzR" "pcaMethods" "Rdisop" "Risa"
[11] "rTANDEM"
CRAN vs Bioconductor packages
> R.version # 3.4.3
# CRAN
> x <- available.packages()
> dim(x)
[1] 12581 17
# Bioconductor Soft
> biocinstallRepos()
BioCsoft
"https://bioconductor.org/packages/3.6/bioc"
BioCann
"https://bioconductor.org/packages/3.6/data/annotation"
BioCexp
"https://bioconductor.org/packages/3.6/data/experiment"
CRAN
"https://cran.rstudio.com/"
> y <- available.packages(repos = biocinstallRepos()[1])
> dim(y)
[1] 1477 17
> intersect(x[, "Package"], y[, "Package"])
character(0)
# Bioconductor Annotation
> dim(available.packages(repos = biocinstallRepos()[2]))
[1] 909 17
# Bioconductor Experiment
> dim(available.packages(repos = biocinstallRepos()[3]))
[1] 326 17
# CRAN + All Bioconductor
> z <- available.packages(repos = biocinstallRepos())
> dim(z)
[1] 15292 17
Downloading Bioconductor package with an old R
When I try to download the GenomicDataCommons package using R 3.4.4 with Bioc 3.6 (the current R version is 3.5.0), it was found it can only install version 1.2.0 instead the latest version 1.4.1.
It does not work by running biocLite("BiocUpgrade") to upgrade Bioc from 3.6 to 3.7.
source("https://bioconductor.org/biocLite.R")
biocLite("BiocUpgrade")
# Error: Bioconductor version 3.6 cannot be upgraded with R version 3.4.4
Analyzing data on CRAN packages
New undocumented function in R 3.4.0: tools::CRAN_package_db()
http://blog.revolutionanalytics.com/2017/05/analyzing-data-on-cran-packages.html
R package location when they are installed by root
/usr/local/lib/R/site-library
Install personal R packages after upgrade R, .libPaths(), Rprofile.site
Scenario: We already have installed many R packages under R 3.1.X in the user's directory. Now we upgrade R to a new version (3.2.X). We like to have these packages available in R 3.2.X.
For Windows OS, refer to R for Windows FAQ
The follow method works on Linux and Windows.
Make sure only one instance of R is running
# Step 1. update R's built-in packages and install them on my personal directory
update.packages(ask=FALSE, checkBuilt = TRUE, repos="http://cran.rstudio.com")
# Step 2. update Bioconductor packages
.libPaths() # The first one is my personal directory
# [1] "/home/brb/R/x86_64-pc-linux-gnu-library/3.2"
# [2] "/usr/local/lib/R/site-library"
# [3] "/usr/lib/R/site-library"
# [4] "/usr/lib/R/library"
Sys.getenv("R_LIBS_USER") # equivalent to .libPaths()[1]
ul <- unlist(strsplit(Sys.getenv("R_LIBS_USER"), "/"))
src <- file.path(paste(ul[1:(length(ul)-1)], collapse="/"), "3.1")
des <- file.path(paste(ul[1:(length(ul)-1)], collapse="/"), "3.2")
pkg <- dir(src, full.names = TRUE)
if (!file.exists(des)) dir.create(des) # If 3.2 subdirectory does not exist yet!
file.copy(pkg, des, overwrite=FALSE, recursive = TRUE)
source("http://www.bioconductor.org/biocLite.R")
biocLite(ask = FALSE)
From Robert Kabacoff (R in Action)
- If you have a customized Rprofile.site file (see appendix B), save a copy outside of R.
- Launch your current version of R and issue the following statements
oldip <- installed.packages()[,1] save(oldip, file="path/installedPackages.Rdata")
where path is a directory outside of R.
- Download and install the newer version of R.
- If you saved a customized version of the Rprofile.site file in step 1, copy it into the new installation.
- Launch the new version of R, and issue the following statements
load("path/installedPackages.Rdata")
newip <- installed.packages()[,1]
for(i in setdiff(oldip, newip))
install.packages(i)
where path is the location specified in step 2.
- Delete the old installation (optional).
This approach will install only packages that are available from the CRAN. It won’t find packages obtained from other locations. In fact, the process will display a list of packages that can’t be installed For example for packages obtained from Bioconductor, use the following method to update packages
source(http://bioconductor.org/biocLite.R) biocLite(PKGNAME)
List vignettes from a package
vignette(package=PACKAGENAME)
List data from a package
data(package=PACKAGENAME)
List installed packages and versions
- http://heuristicandrew.blogspot.com/2015/06/list-of-user-installed-r-packages-and.html
- checkpoint package
ip <- as.data.frame(installed.packages()[,c(1,3:4)]) rownames(ip) <- NULL unique(ip$Priority) # [1] <NA> base recommended # Levels: base recommended ip <- ip[is.na(ip$Priority),1:2,drop=FALSE] print(ip, row.names=FALSE)
Query the names of outdated packages
psi <- packageStatus()$inst subset(psi, Status == "upgrade", drop = FALSE) # Package LibPath Version Priority Depends # RcppArmadillo RcppArmadillo C:/Users/brb/Documents/R/win-library/3.2 0.5.100.1.0 <NA> <NA> # Matrix Matrix C:/Program Files/R/R-3.2.0/library 1.2-0 recommended R (>= 2.15.2), methods # Imports LinkingTo Suggests # RcppArmadillo Rcpp (>= 0.11.0) Rcpp RUnit, Matrix, pkgKitten # Matrix graphics, grid, stats, utils, lattice <NA> expm, MASS # Enhances License License_is_FOSS License_restricts_use OS_type MD5sum # RcppArmadillo <NA> GPL (>= 2) <NA> <NA> <NA> <NA> # Matrix MatrixModels, graph, SparseM, sfsmisc GPL (>= 2) <NA> <NA> <NA> <NA> # NeedsCompilation Built Status # RcppArmadillo yes 3.2.0 upgrade # Matrix yes 3.2.0 upgrade
The above output does not show the package version from the latest packages on CRAN. So the following snippet does that.
psi <- packageStatus()$inst
pl <- unname(psi$Package[psi$Status == "upgrade"]) # List package names
ap <- as.data.frame(available.packages()[, c(1,2,3)], stringsAsFactors = FALSE)
out <- cbind(subset(psi, Status == "upgrade")[, c("Package", "Version")], ap[match(pl, ap$Package), "Version"])
colnames(out)[2:3] <- c("OldVersion", "NewVersion")
rownames(out) <- NULL
out
# Package OldVersion NewVersion
# 1 RcppArmadillo 0.5.100.1.0 0.5.200.1.0
# 2 Matrix 1.2-0 1.2-1
To consider also the packages from Bioconductor, we have the following code. Note that "3.1" means the Bioconductor version and "3.2" is the R version. See Bioconductor release versions page.
psic <- packageStatus(repos = c(contrib.url(getOption("repos")),
"http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2",
"http://www.bioconductor.org/packages/3.1/data/annotation/bin/windows/contrib/3.2"))$inst
subset(psic, Status == "upgrade", drop = FALSE)
pl <- unname(psic$Package[psic$Status == "upgrade"])
ap <- as.data.frame(available.packages(c(contrib.url(getOption("repos")),
"http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2",
"http://www.bioconductor.org/packages/3.1/data/annotation/bin/windows/contrib/3.2"))[, c(1:3)],
stringAsFactors = FALSE)
out <- cbind(subset(psic, Status == "upgrade")[, c("Package", "Version")], ap[match(pl, ap$Package), "Version"])
colnames(out)[2:3] <- c("OldVersion", "NewVersion")
rownames(out) <- NULL
out
# Package OldVersion NewVersion
# 1 limma 3.24.5 3.24.9
# 2 RcppArmadillo 0.5.100.1.0 0.5.200.1.0
# 3 Matrix 1.2-0 1.2-1
Searching for packages in CRAN
- pkgsearch package - Search R packages on CRAN
- Fishing for packages in CRAN
- CRAN now has 10,000 R packages. Here's how to find the ones you need
- Searching for R packages, packagefinder package
library(packagefinder) findPackage("survival") # 272 out of 13256 CRAN packages found in 5 seconds findPackage("follic") # No results found. # Actually 'follic' comes from randomForestSRC package # https://www.rdocumentation.org/packages/randomForestSRC/versions/2.7.0The result is shown in an html format with columns of SCORE, NAME, DESC_SHORT, DOWNL_TOTAL & GO.
METACRAN - Search and browse all CRAN/R packages
- Source code on https://github.com/metacran. The 'PACKAGES' file is updated regularly to Github.
- Announcement on R/mailing list
- Author's homepage on http://gaborcsardi.org/.
cranly visualisations and summaries for R packages
Exploring R packages with cranly
Query top downloaded packages
- cranlogs package - Download Logs from the RStudio CRAN Mirror
- http://blog.revolutionanalytics.com/2015/06/working-with-the-rstudio-cran-logs.html
Would you like to use a personal library instead?
Some posts from internet
- Setting R_LIBS & avoiding “Would you like to use a personal library instead?”. Note: I try to create ~/.Renviron to add my personal folder in it. But update.packages() still asks me if I like to use a personal library instead (tested on Ubuntu + R 3.4).
- automatically create personal library in R. Using suppressUpdates + specify lib in biocLite() or update.packages(Sys.getenv("R_LIBS_USER"), ask = F)
# create local user library path (not present by default) dir.create(path = Sys.getenv("R_LIBS_USER"), showWarnings = FALSE, recursive = TRUE) # install to local user library path install.packages(p, lib = Sys.getenv("R_LIBS_USER"), repos = "https://cran.rstudio.com/") # Bioconductor version biocLite(p, suppressUpdates = TRUE, lib = Sys.getenv("R_LIBS_USER"))
The problem can happen if the R was installed to the C:\Program Files\R folder by users but then some main packages want to be upgraded. R will always pops a message 'Would you like to use a personal library instead?'.
To suppress the message and use the personal library always,
- Run R as administrator. If you do that, main packages can be upgraded from C:\Program Files\R\R-X.Y.Z\library folder.
- Writable R package directory cannot be found and a this. A solution here is to change the security of the R library folder so the user has a full control on the folder.
- Does R run under Windows Vista/7/8/Server 2008? There are 3 ways to get around the issue.
- I don’t have permission to write to the R-3.3.2\library directory
Actually the following hints will help us to create a convenient function UpdateMainLibrary() which will install updated main packages in the user's Documents directory without a warning dialog.
- .libPaths() only returns 1 string "C:/Program Files/R/R-x.y.z/library" on the machines that does not have this problem
- .libPaths() returns two strings "C:/Users/USERNAME/Documents/R/win-library/x.y" & "C:/Program Files/R/R-x.y.z/library" on machines with the problem.
UpdateMainLibrary <- function() {
# Update main/site packages
# The function is used to fix the problem 'Would you like to use a personal library instead?'
if (length(.libPaths()) == 1) return()
ind_mloc <- grep("Program", .libPaths()) # main library e.g. 2
ind_ploc <- grep("Documents", .libPaths()) # personal library e.g. 1
if (length(ind_mloc) > 0L && length(ind_ploc) > 0L)
# search outdated main packages
old_mloc <- ! old.packages(.libPaths()[ind_mloc])[, "Package"] %in%
installed.packages(.libPaths()[ind_ploc])[, "Package"]
oldpac <- old.packages(.libPaths()[ind_mloc])[old_mloc, "Package"]
if (length(oldpac) > 0L)
install.packages(oldpac, .libPaths()[ind_ploc])
}
On Linux,
> update.packages()
...
The downloaded source packages are in
‘/tmp/RtmpBrYccd/downloaded_packages’
Warning in install.packages(update[instlib == l, "Package"], l, contriburl = contriburl, :
'lib = "/opt/R/3.5.0/lib/R/library"' is not writable
Would you like to use a personal library instead? (yes/No/cancel) yes
...
> system("ls -lt /home/brb/R/x86_64-pc-linux-gnu-library/3.5 | head")
total 224
drwxrwxr-x 9 brb brb 4096 Oct 3 09:30 survival
drwxrwxr-x 9 brb brb 4096 Oct 3 09:29 mgcv
drwxrwxr-x 10 brb brb 4096 Oct 3 09:29 MASS
drwxrwxr-x 9 brb brb 4096 Oct 3 09:29 foreign
# So new versions of survival, mgc, MASS, foreign are installed in the personal directory
# The update.packages() will issue warnings if we try to run it again.
# It's OK to ignore these warnings.
> update.packages()
Warning: package 'foreign' in library '/opt/R/3.5.0/lib/R/library' will not be updated
Warning: package 'MASS' in library '/opt/R/3.5.0/lib/R/library' will not be updated
Warning: package 'mgcv' in library '/opt/R/3.5.0/lib/R/library' will not be updated
Warning: package 'survival' in library '/opt/R/3.5.0/lib/R/library' will not be updated
installation path not writeable from running biocLite()
When I ran biocLite() to install a new package, I got a message (the Bioc packages are installed successfully anyway)
... * DONE (curatedOvarianData) The downloaded source packages are in ‘/tmp/RtmpHxnH2K/downloaded_packages’ installation path not writeable, unable to update packages: rgl, rJava, codetools, foreign, lattice, MASS, spatial, survival
However, if I uses install.package() it can update the package
> packageVersion("survival")
[1] ‘2.42.3’
> update.packages("survival") # Not working though no error message
> packageVersion("survival")
[1] ‘2.42.3’
> install.packages("survival")
Installing package into ‘/home/brb/R/x86_64-pc-linux-gnu-library/3.4’
...
* DONE (survival)
The downloaded source packages are in
‘/tmp/RtmpHxnH2K/downloaded_packages’
> packageVersion("survival")
[1] ‘2.42.6’
> library(survival)
> sessionInfo() # show survival package 2.42-6 was attached
It makes sense to always use personal directory when we install packages. See .libPaths().
Warning: cannot remove prior installation of package
Instance 1.
# Install the latest hgu133plus2cdf package
# Remove/Uninstall hgu133plus2.db package
# Put/Install an old version of IRanges (eg version 1.18.2 while currently it is version 1.18.3)
# Test on R 3.0.1
library(hgu133plus2cdf) # hgu133pluscdf does not depend or import IRanges
source("http://bioconductor.org/biocLite.R")
biocLite("hgu133plus2.db", ask=FALSE) # hgu133plus2.db imports IRanges
# Warning:cannot remove prior installation of package 'IRanges'
# Open Windows Explorer and check IRanges folder. Only see libs subfolder.
Note:
- In the above example, all packages were installed under C:\Program Files\R\R-3.0.1\library\.
- In another instance where I cannot reproduce the problem, new R packages were installed under C:\Users\xxx\Documents\R\win-library\3.0\. The different thing is IRanges package CAN be updated but if I use packageVersion("IRanges") command in R, it still shows the old version.
- The above were tested on a desktop.
Instance 2.
# On a fresh R 3.2.0, I install Bioconductor's depPkgTools & lumi packages. Then I close R, re-open it,
# and install depPkgTools package again.
> source("http://bioconductor.org/biocLite.R")
Bioconductor version 3.1 (BiocInstaller 1.18.2), ?biocLite for help
> biocLite("pkgDepTools")
BioC_mirror: http://bioconductor.org
Using Bioconductor version 3.1 (BiocInstaller 1.18.2), R version 3.2.0.
Installing package(s) ‘pkgDepTools’
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/pkgDepTools_1.34.0.zip'
Content type 'application/zip' length 390579 bytes (381 KB)
downloaded 381 KB
package ‘pkgDepTools’ successfully unpacked and MD5 sums checked
Warning: cannot remove prior installation of package ‘pkgDepTools’
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpYd2l7i\downloaded_packages
> library(pkgDepTools)
Error in library(pkgDepTools) : there is no package called ‘pkgDepTools’
The pkgDepTools library folder appears in C:\Users\brb\Documents\R\win-library\3.2, but it is empty. The weird thing is if I try the above steps again, I cannot reproduce the problem.
Warning: Unable to move temporary installation
The problem seems to happen only on virtual machines (Virtualbox).
- Warning: unable to move temporary installation `C:\Users\brb\Documents\R\win-library\3.0\fileed8270978f5\quadprog` to `C:\Users\brb\Documents\R\win-library\3.0\quadprog` when I try to run 'install.packages("forecast").
- Warning: unable to move temporary installation ‘C:\Users\brb\Documents\R\win-library\3.2\file5e0104b5b49\plyr’ to ‘C:\Users\brb\Documents\R\win-library\3.2\plyr’ when I try to run 'biocLite("lumi")'. The other dependency packages look fine although I am not sure if any unknown problem can happen (it does, see below).
Here is a note of my trouble shooting.
- If I try to ignore the warning and load the lumi package. I will get an error.
- If I try to run biocLite("lumi") again, it will only download & install lumi without checking missing 'plyr' package. Therefore, when I try to load the lumi package, it will give me an error again.
- Even I install the plyr package manually, library(lumi) gives another error - missing mclust package.
> biocLite("lumi")
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/BiocInstaller_1.18.2.zip'
Content type 'application/zip' length 114097 bytes (111 KB)
downloaded 111 KB
...
package ‘lumi’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages
Old packages: 'BiocParallel', 'Biostrings', 'caret', 'DESeq2', 'gdata', 'GenomicFeatures', 'gplots', 'Hmisc', 'Rcpp', 'RcppArmadillo', 'rgl',
'stringr'
Update all/some/none? [a/s/n]: a
also installing the dependencies ‘Rsamtools’, ‘GenomicAlignments’, ‘plyr’, ‘rtracklayer’, ‘gridExtra’, ‘stringi’, ‘magrittr’
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/Rsamtools_1.20.1.zip'
Content type 'application/zip' length 8138197 bytes (7.8 MB)
downloaded 7.8 MB
...
package ‘Rsamtools’ successfully unpacked and MD5 sums checked
package ‘GenomicAlignments’ successfully unpacked and MD5 sums checked
package ‘plyr’ successfully unpacked and MD5 sums checked
Warning: unable to move temporary installation ‘C:\Users\brb\Documents\R\win-library\3.2\file5e0104b5b49\plyr’
to ‘C:\Users\brb\Documents\R\win-library\3.2\plyr’
package ‘rtracklayer’ successfully unpacked and MD5 sums checked
package ‘gridExtra’ successfully unpacked and MD5 sums checked
package ‘stringi’ successfully unpacked and MD5 sums checked
package ‘magrittr’ successfully unpacked and MD5 sums checked
package ‘BiocParallel’ successfully unpacked and MD5 sums checked
package ‘Biostrings’ successfully unpacked and MD5 sums checked
Warning: cannot remove prior installation of package ‘Biostrings’
package ‘caret’ successfully unpacked and MD5 sums checked
package ‘DESeq2’ successfully unpacked and MD5 sums checked
package ‘gdata’ successfully unpacked and MD5 sums checked
package ‘GenomicFeatures’ successfully unpacked and MD5 sums checked
package ‘gplots’ successfully unpacked and MD5 sums checked
package ‘Hmisc’ successfully unpacked and MD5 sums checked
package ‘Rcpp’ successfully unpacked and MD5 sums checked
package ‘RcppArmadillo’ successfully unpacked and MD5 sums checked
package ‘rgl’ successfully unpacked and MD5 sums checked
package ‘stringr’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages
> library(lumi)
Error in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vI[[i]]) :
there is no package called ‘plyr’
Error: package or namespace load failed for ‘lumi’
> search()
[1] ".GlobalEnv" "package:BiocInstaller" "package:Biobase" "package:BiocGenerics" "package:parallel" "package:stats"
[7] "package:graphics" "package:grDevices" "package:utils" "package:datasets" "package:methods" "Autoloads"
[13] "package:base"
> biocLite("lumi")
BioC_mirror: http://bioconductor.org
Using Bioconductor version 3.1 (BiocInstaller 1.18.2), R version 3.2.0.
Installing package(s) ‘lumi’
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/lumi_2.20.1.zip'
Content type 'application/zip' length 18185326 bytes (17.3 MB)
downloaded 17.3 MB
package ‘lumi’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages
> search()
[1] ".GlobalEnv" "package:BiocInstaller" "package:Biobase" "package:BiocGenerics" "package:parallel" "package:stats"
[7] "package:graphics" "package:grDevices" "package:utils" "package:datasets" "package:methods" "Autoloads"
[13] "package:base"
> library(lumi)
Error in loadNamespace(i, c(lib.loc, .libPaths()), versionCheck = vI[[i]]) :
there is no package called ‘plyr’
Error: package or namespace load failed for ‘lumi’
> biocLite("plyr")
BioC_mirror: http://bioconductor.org
Using Bioconductor version 3.1 (BiocInstaller 1.18.2), R version 3.2.0.
Installing package(s) ‘plyr’
trying URL 'http://cran.rstudio.com/bin/windows/contrib/3.2/plyr_1.8.2.zip'
Content type 'application/zip' length 1128621 bytes (1.1 MB)
downloaded 1.1 MB
package ‘plyr’ successfully unpacked and MD5 sums checked
The downloaded binary packages are in
C:\Users\brb\AppData\Local\Temp\RtmpyUjsJD\downloaded_packages
> library(lumi)
Error in loadNamespace(j <- i[[1L]], c(lib.loc, .libPaths()), versionCheck = vI[[j]]) :
there is no package called ‘mclust’
Error: package or namespace load failed for ‘lumi’
> ?biocLite
Warning messages:
1: In read.dcf(file.path(p, "DESCRIPTION"), c("Package", "Version")) :
cannot open compressed file 'C:/Users/brb/Documents/R/win-library/3.2/Biostrings/DESCRIPTION', probable reason 'No such file or directory'
2: In find.package(if (is.null(package)) loadedNamespaces() else package, :
there is no package called ‘Biostrings’
> library(lumi)
Error in loadNamespace(j <- i[[1L]], c(lib.loc, .libPaths()), versionCheck = vI[[j]]) :
there is no package called ‘mclust’
In addition: Warning messages:
1: In read.dcf(file.path(p, "DESCRIPTION"), c("Package", "Version")) :
cannot open compressed file 'C:/Users/brb/Documents/R/win-library/3.2/Biostrings/DESCRIPTION', probable reason 'No such file or directory'
2: In find.package(if (is.null(package)) loadedNamespaces() else package, :
there is no package called ‘Biostrings’
Error: package or namespace load failed for ‘lumi’
Other people also have the similar problem. The possible cause is the virus scanner locks the file and R cannot move them.
Some possible solutions:
- Delete ALL folders under R/library (e.g. C:/Progra~1/R/R-3.2.0/library) folder and install the main package again using install.packages() or biocLite().
- For specific package like 'lumi' from Bioconductor, we can find out all dependency packages and then install them one by one.
- Find out and install the top level package which misses dependency packages.
- This is based on the fact that install.packages() or biocLite() sometimes just checks & installs the 'Depends' and 'Imports' packages and won't install all packages recursively
- we can do a small experiment by removing a package which is not directly dependent/imported by another package; e.g. 'iterators' is not dependent/imported by 'glment' directly but indirectly. So if we run remove.packages("iterators"); install.packages("glmnet"), then the 'iterator' package is still missing.
- A real example is if the missing packages are 'Biostrings', 'limma', 'mclust' (these are packages that 'minfi' directly depends/imports although they should be installed when I run biocLite("lumi") command), then I should just run the command remove.packages("minfi"); biocLite("minfi"). If we just run biocLite("lumi") or biocLite("methylumi"), the missing packages won't be installed.
Error in download.file(url, destfile, method, mode = "wb", ...)
HTTP status was '404 Not Found'
Tested on an existing R-3.2.0 session. Note that VariantAnnotation 1.14.4 was just uploaded to Bioc.
> biocLite("COSMIC.67")
BioC_mirror: http://bioconductor.org
Using Bioconductor version 3.1 (BiocInstaller 1.18.3), R version 3.2.0.
Installing package(s) ‘COSMIC.67’
also installing the dependency ‘VariantAnnotation’
trying URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/VariantAnnotation_1.14.3.zip'
Error in download.file(url, destfile, method, mode = "wb", ...) :
cannot open URL 'http://bioconductor.org/packages/3.1/bioc/bin/windows/contrib/3.2/VariantAnnotation_1.14.3.zip'
In addition: Warning message:
In download.file(url, destfile, method, mode = "wb", ...) :
cannot open: HTTP status was '404 Not Found'
Warning in download.packages(pkgs, destdir = tmpd, available = available, :
download of package ‘VariantAnnotation’ failed
installing the source package ‘COSMIC.67’
trying URL 'http://bioconductor.org/packages/3.1/data/experiment/src/contrib/COSMIC.67_1.4.0.tar.gz'
Content type 'application/x-gzip' length 40999037 bytes (39.1 MB)
However, when I tested on a new R-3.2.0 (just installed in VM), the COSMIC package installation is successful. That VariantAnnotation version 1.14.4 was installed (this version was just updated today from Bioconductor).
The cause of the error is the available.package() function will read the rds file first from cache in a tempdir (C:\Users\XXXX\AppData\Local\Temp\RtmpYYYYYY). See lines 51-55 of <packages.R>.
dest <- file.path(tempdir(),
paste0("repos_", URLencode(repos, TRUE), ".rds"))
if(file.exists(dest)) {
res0 <- readRDS(dest)
} else {
...
}
Since my R was opened 1 week ago, the rds file it reads today contains old information. Note that Bioconductor does not hold the source code or binary code for the old version of packages. This explains why biocLite() function broke. When I restart R, the original problem is gone.
If we look at the source code of available.packages(), we will see we could use cacheOK option in download.file() function.
download.file(url, destfile, method, cacheOK = FALSE, quiet = TRUE, mode ="wb")
Another case: Error in download.file(url, destfile, method, mode = "wb", ...)
> install.packages("quantreg")
There is a binary version available but the source version is later:
binary source needs_compilation
quantreg 5.33 5.34 TRUE
Do you want to install from sources the package which needs compilation?
y/n: n
trying URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.4/quantreg_5.33.tgz'
Warning in install.packages :
cannot open URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.4/quantreg_5.33.tgz': HTTP status was '404 Not Found'
Error in download.file(url, destfile, method, mode = "wb", ...) :
cannot open URL 'https://cran.rstudio.com/bin/macosx/el-capitan/contrib/3.4/quantreg_5.33.tgz'
Warning in install.packages :
download of package ‘quantreg’ failed
It seems the binary package cannot be found on the mirror. So the solution here is to download the package from the R main server. Note that after I have successfully installed the binary package from the main R server, I remove the package in R and try to install the binary package from rstudio.com server agin and it works this time.
> install.packages("quantreg", repos = "https://cran.r-project.org")
trying URL 'https://cran.r-project.org/bin/macosx/el-capitan/contrib/3.4/quantreg_5.34.tgz'
Content type 'application/x-gzip' length 1863561 bytes (1.8 MB)
==================================================
downloaded 1.8 MB
Another case: Error in download.file() on Windows 7
For some reason, IE 8 cannot interpret https://ftp.ncbi.nlm.nih.gov though it understands ftp://ftp.ncbi.nlm.nih.gov.
This is tested using R 3.4.3.
> download.file("https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", "test.soft.gz")
trying URL 'https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz'
Error in download.file("https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", :
cannot open URL 'https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz'
In addition: Warning message:
In download.file("https://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", :
InternetOpenUrl failed: 'An error occurred in the secure channel support'
> download.file("ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz", "test.soft.gz")
trying URL 'ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE7nnn/GSE7848/soft/GSE7848_family.soft.gz'
downloaded 9.1 MB
Error in unloadNamespace(package)
> d3heatmap(mtcars, scale = "column", colors = "Blues")
Error: 'col_numeric' is not an exported object from 'namespace:scales'
> packageVersion("scales")
[1] ‘0.2.5’
> library(scales)
Error in unloadNamespace(package) :
namespace ‘scales’ is imported by ‘ggplot2’ so cannot be unloaded
In addition: Warning message:
package ‘scales’ was built under R version 3.2.1
Error in library(scales) :
Package ‘scales’ version 0.2.4 cannot be unloaded
> search()
[1] ".GlobalEnv" "package:d3heatmap" "package:ggplot2"
[4] "package:microbenchmark" "package:COSMIC.67" "package:BiocInstaller"
[7] "package:stats" "package:graphics" "package:grDevices"
[10] "package:utils" "package:datasets" "package:methods"
[13] "Autoloads" "package:base"
If I open a new R session, the above error will not happen!
The problem occurred because the 'scales' package version required by the d3heatmap package/function is old. See this post. And when I upgraded the 'scales' package, it was locked by the package was imported by the ggplot2 package.
Unload a package
See an example below.
require(splines) detach(package:splines, unload=TRUE)
New R packages as reported by CRANberries
http://blog.revolutionanalytics.com/2015/07/mranspackages-spotlight.html
#----------------------------
# SCRAPE CRANBERRIES FILES TO COUNT NEW PACKAGES AND PLOT
#
library(ggplot2)
# Build a vextor of the directories of interest
year <- c("2013","2014","2015")
month <- c("01","02","03","04","05","06","07","08","09","10","11","12")
span <-c(rep(month,2),month[1:7])
dir <- "http://dirk.eddelbuettel.com/cranberries"
url2013 <- file.path(dir,"2013",month)
url2014 <- file.path(dir,"2014",month)
url2015 <- file.path(dir,"2015",month[1:7])
url <- c(url2013,url2014,url2015)
# Read each directory and count the new packages
new_p <- vector()
for(i in url){
raw.data <- readLines(i)
new_p[i] <- length(grep("New package",raw.data,value=TRUE))
}
# Plot
time <- seq(as.Date("2013-01-01"), as.Date("2015-07-01"), by="months")
new_pkgs <- data.frame(time,new_p)
ggplot(new_pkgs, aes(time,y=new_p)) +
geom_line() + xlab("") + ylab("Number of new packages") +
geom_smooth(method='lm') + ggtitle("New R packages as reported by CRANberries")
Top new packages in 2015
- 2015 R packages roundup by CHRISTOPH SAFFERLING
- R trends in 2015 by MAX GORDON
Speeding up package installation
- http://blog.jumpingrivers.com/posts/2017/speed_package_installation/
- (Much) Faster Package (Re-)Installation via Caching
- (Much) Faster Package (Re-)Installation via Caching, part 2
package ‘XXX’ was installed by an R version with different internals
it needs to be reinstalled for use with this R version. The problem seems to be specific to R 3.5.1 in Ubuntu 16.04. I got this message when I try to install the keras and tidyverse packages. The 'XXX' package includes nlme for installing "tidyverse" and Matrix for installing "reticulate". I have already logged in as root. I need to manually install these packages again though it seems I did not see a version change for these packages.
R package dependencies
- Package tools' functions package.dependencies(), pkgDepends(), etc are deprecated now, mostly in favor of package_dependencies() which is both more flexible and efficient. See R 3.3.0 News.
Depends, Imports, Suggests, Enhances, LinkingTo
See Writing R Extensions and install.packages().
- Depends: list of package names which this package depends on. Those packages will be attached (so it is better to use Imports instead of Depends as much as you can) before the current package when library or require is called. The ‘Depends’ field can also specify a dependence on a certain version of R.
- Imports: lists packages whose namespaces are imported from (as specified in the NAMESPACE file) but which do not need to be attached.
- Suggests: lists packages that are not necessarily needed. This includes packages used only in examples, tests or vignettes, and packages loaded in the body of functions.
- Enhances: lists packages “enhanced” by the package at hand, e.g., by providing methods for classes from these packages, or ways to handle objects from these packages.
- LinkingTo: A package that wishes to make use of header files in other packages needs to declare them as a comma-separated list in the field ‘LinkingTo’ in the DESCRIPTION file.
Bioconductor's pkgDepTools package
The is an example of querying the dependencies of the notorious 'lumi' package which often broke the installation script. I am using R 3.2.0 and Bioconductor 3.1.
The getInstallOrder function is useful to get a list of all (recursive) dependency packages.
source("http://bioconductor.org/biocLite.R")
if (!require(pkgDepTools)) {
biocLite("pkgDepTools", ask = FALSE)
library(pkgDepTools)
}
MkPlot <- FALSE
library(BiocInstaller)
biocUrl <- biocinstallRepos()["BioCsoft"]
biocDeps <- makeDepGraph(biocUrl, type="source", dosize=FALSE) # pkgDepTools defines its makeDepGraph()
PKG <- "lumi"
if (MkPlot) {
if (!require(Biobase)) {
biocLite("Biobase", ask = FALSE)
library(Biobase)
}
if (!require(Rgraphviz)) {
biocLite("Rgraphviz", ask = FALSE)
library(Rgraphviz)
}
categoryNodes <- c(PKG, names(acc(biocDeps, PKG)[[1]]))
categoryGraph <- subGraph(categoryNodes, biocDeps)
nn <- makeNodeAttrs(categoryGraph, shape="ellipse")
plot(categoryGraph, nodeAttrs=nn) # Complete but plot is too complicated & font is too small.
}
system.time(allDeps <- makeDepGraph(biocinstallRepos(), type="source",
keep.builtin=TRUE, dosize=FALSE)) # takes a little while
# user system elapsed
# 175.737 10.994 186.875
# Warning messages:
# 1: In .local(from, to, graph) : edges replaced: ‘SNPRelate|gdsfmt’
# 2: In .local(from, to, graph) :
# edges replaced: ‘RCurl|methods’, ‘NA|bitops’
# When needed.only=TRUE, only those dependencies not currently installed are included in the list.
x1 <- sort(getInstallOrder(PKG, allDeps, needed.only=TRUE)$packages); x1
[1] "affy" "affyio"
[3] "annotate" "AnnotationDbi"
[5] "base64" "beanplot"
[7] "Biobase" "BiocParallel"
[9] "biomaRt" "Biostrings"
[11] "bitops" "bumphunter"
[13] "colorspace" "DBI"
[15] "dichromat" "digest"
[17] "doRNG" "FDb.InfiniumMethylation.hg19"
[19] "foreach" "futile.logger"
[21] "futile.options" "genefilter"
[23] "GenomeInfoDb" "GenomicAlignments"
[25] "GenomicFeatures" "GenomicRanges"
[27] "GEOquery" "ggplot2"
[29] "gtable" "illuminaio"
[31] "IRanges" "iterators"
[33] "labeling" "lambda.r"
[35] "limma" "locfit"
[37] "lumi" "magrittr"
[39] "matrixStats" "mclust"
[41] "methylumi" "minfi"
[43] "multtest" "munsell"
[45] "nleqslv" "nor1mix"
[47] "org.Hs.eg.db" "pkgmaker"
[49] "plyr" "preprocessCore"
[51] "proto" "quadprog"
[53] "RColorBrewer" "Rcpp"
[55] "RCurl" "registry"
[57] "reshape" "reshape2"
[59] "rngtools" "Rsamtools"
[61] "RSQLite" "rtracklayer"
[63] "S4Vectors" "scales"
[65] "siggenes" "snow"
[67] "stringi" "stringr"
[69] "TxDb.Hsapiens.UCSC.hg19.knownGene" "XML"
[71] "xtable" "XVector"
[73] "zlibbioc"
# When needed.only=FALSE the complete list of dependencies is given regardless of the set of currently installed packages.
x2 <- sort(getInstallOrder(PKG, allDeps, needed.only=FALSE)$packages); x2
[1] "affy" "affyio" "annotate"
[4] "AnnotationDbi" "base64" "beanplot"
[7] "Biobase" "BiocGenerics" "BiocInstaller"
[10] "BiocParallel" "biomaRt" "Biostrings"
[13] "bitops" "bumphunter" "codetools"
[16] "colorspace" "DBI" "dichromat"
[19] "digest" "doRNG" "FDb.InfiniumMethylation.hg19"
[22] "foreach" "futile.logger" "futile.options"
[25] "genefilter" "GenomeInfoDb" "GenomicAlignments"
[28] "GenomicFeatures" "GenomicRanges" "GEOquery"
[31] "ggplot2" "graphics" "grDevices"
[34] "grid" "gtable" "illuminaio"
[37] "IRanges" "iterators" "KernSmooth"
[40] "labeling" "lambda.r" "lattice"
[43] "limma" "locfit" "lumi"
[46] "magrittr" "MASS" "Matrix"
[49] "matrixStats" "mclust" "methods"
[52] "methylumi" "mgcv" "minfi"
[55] "multtest" "munsell" "nleqslv"
[58] "nlme" "nor1mix" "org.Hs.eg.db"
[61] "parallel" "pkgmaker" "plyr"
[64] "preprocessCore" "proto" "quadprog"
[67] "RColorBrewer" "Rcpp" "RCurl"
[70] "registry" "reshape" "reshape2"
[73] "rngtools" "Rsamtools" "RSQLite"
[76] "rtracklayer" "S4Vectors" "scales"
[79] "siggenes" "snow" "splines"
[82] "stats" "stats4" "stringi"
[85] "stringr" "survival" "tools"
[88] "TxDb.Hsapiens.UCSC.hg19.knownGene" "utils" "XML"
[91] "xtable" "XVector" "zlibbioc"
> sort(setdiff(x2, x1)) # Not all R's base packages are included; e.g. 'base', 'boot', ...
[1] "BiocGenerics" "BiocInstaller" "codetools" "graphics" "grDevices"
[6] "grid" "KernSmooth" "lattice" "MASS" "Matrix"
[11] "methods" "mgcv" "nlme" "parallel" "splines"
[16] "stats" "stats4" "survival" "tools" "utils"
Bioconductor BiocPkgTools
Collection of simple tools for learning about Bioc Packages
Overview of BiocPkgTools & Dependency graphs
miniCRAN package
miniCRAN package can be used to identify package dependencies or create a local CRAN repository. It can be used on repositories other than CRAN, such as Bioconductor.
- http://blog.revolutionanalytics.com/2014/07/dependencies-of-popular-r-packages.html
- http://www.r-bloggers.com/introducing-minicran-an-r-package-to-create-a-private-cran-repository/
- http://www.magesblog.com/2014/09/managing-r-package-dependencies.html
- Using miniCRAN in Azure ML
- developing internal CRAN Repositories
Before we go into R, we need to install some packages from Ubuntu terminal. See here.
# Consider glmnet package (today is 4/29/2015)
# Version: 2.0-2
# Depends: Matrix (≥ 1.0-6), utils, foreach
# Suggests: survival, knitr, lars
if (!require("miniCRAN")) {
install.packages("miniCRAN", dependencies = TRUE, repos="http://cran.rstudio.com") # include 'igraph' in Suggests.
library(miniCRAN)
}
if (!"igraph" %in% installed.packages()[,1]) install.packages("igraph")
tags <- "glmnet"
pkgDep(tags, suggests=TRUE, enhances=TRUE) # same as pkgDep(tags)
# [1] "glmnet" "Matrix" "foreach" "codetools" "iterators" "lattice" "evaluate" "digest"
# [9] "formatR" "highr" "markdown" "stringr" "yaml" "mime" "survival" "knitr"
# [17] "lars"
dg <- makeDepGraph(tags, suggests=TRUE, enhances=TRUE) # miniCRAN defines its makeDepGraph()
plot(dg, legendPosition = c(-1, 1), vertex.size=20)
We can also display the dependence for a package from the Bioconductor repository.
tags <- "DESeq2"
# Depends S4Vectors, IRanges, GenomicRanges, Rcpp (>= 0.10.1), RcppArmadillo (>= 0.3.4.4)
# Imports BiocGenerics(>= 0.7.5), Biobase, BiocParallel, genefilter, methods, locfit, geneplotter, ggplot2, Hmisc
# Suggests RUnit, gplots, knitr, RColorBrewer, BiocStyle, airway,\npasilla (>= 0.2.10), DESeq, vsn
# LinkingTo Rcpp, RcppArmadillo
index <- function(url, type="source", filters=NULL, head=5, cols=c("Package", "Version")){
contribUrl <- contrib.url(url, type=type)
available.packages(contribUrl, type=type, filters=filters)
}
bioc <- local({
env <- new.env()
on.exit(rm(env))
evalq(source("http://bioconductor.org/biocLite.R", local=TRUE), env)
biocinstallRepos() # return URLs
})
bioc
# BioCsoft
# "http://bioconductor.org/packages/3.0/bioc"
# BioCann
# "http://bioconductor.org/packages/3.0/data/annotation"
# BioCexp
# "http://bioconductor.org/packages/3.0/data/experiment"
# BioCextra
# "http://bioconductor.org/packages/3.0/extra"
# CRAN
# "http://cran.fhcrc.org"
# CRANextra
# "http://www.stats.ox.ac.uk/pub/RWin"
str(index(bioc["BioCsoft"])) # similar to cranJuly2014 object
system.time(dg <- makeDepGraph(tags, suggests=TRUE, enhances=TRUE, availPkgs = index(bioc["BioCsoft"]))) # Very quick!
plot(dg, legendPosition = c(-1, 1), vertex.size=20)
The dependencies of GenomicFeature and GenomicAlignments are more complicated. So we turn the 'suggests' option to FALSE.
tags <- "GenomicAlignments" dg <- makeDepGraph(tags, suggests=FALSE, enhances=FALSE, availPkgs = index(bioc["BioCsoft"])) plot(dg, legendPosition = c(-1, 1), vertex.size=20)
MRAN (CRAN only)
cranly
Reverse dependence
Install packages offline
http://www.mango-solutions.com/wp/2017/05/installing-packages-without-internet/
Install a packages locally and its dependencies
It's impossible to install the dependencies if you want to install a package locally. See Windows-GUI: "Install Packages from local zip files" and dependencies
Create a new R package, namespace, documentation
- http://cran.r-project.org/doc/contrib/Leisch-CreatingPackages.pdf (highly recommend)
- https://stat.ethz.ch/pipermail/r-devel/2013-July/066975.html
- Benefit of import in a namespace
- This youtube video from Tyler Rinker teaches how to use RStudio to develop an R package and also use Git to do version control. Very useful!
- Developing R packages by Jeff Leek in Johns Hopkins University.
- R packages book by Hadley Wickham.
- R package primer a minimal tutorial from Karl Broman.
- How to make and share an R package in 3 steps (6/14/2017)
R package depends vs imports
- http://stackoverflow.com/questions/8637993/better-explanation-of-when-to-use-imports-depends
- http://stackoverflow.com/questions/9893791/imports-and-depends
- https://stat.ethz.ch/pipermail/r-devel/2013-August/067082.html
In the namespace era Depends is never really needed. All modern packages have no technical need for Depends anymore. Loosely speaking the only purpose of Depends today is to expose other package's functions to the user without re-exporting them.
load = functions exported in myPkg are available to interested parties as myPkg::foo or via direct imports - essentially this means the package can now be used
attach = the namespace (and thus all exported functions) is attached to the search path - the only effect is that you have now added the exported functions to the global pool of functions - sort of like dumping them in the workspace (for all practical purposes, not technically)
import a function into a package = make sure that this function works in my package regardless of the search path (so I can write fn1 instead of pkg1::fn1 and still know it will come from pkg1 and not someone's workspace or other package that chose the same name)
The distinction is between "loading" and "attaching" a package. Loading it (which would be done if you had MASS::loglm, or imported it) guarantees that the package is initialized and in memory, but doesn't make it visible to the user without the explicit MASS:: prefix. Attaching it first loads it, then modifies the user's search list so the user can see it.
Loading is less intrusive, so it's preferred over attaching. Both library() and require() would attach it.
R package suggests
stringr has suggested htmlwidgets. An error will come out if the suggested packages are not available.
> library(stringr)
> str_view(c("abc", "a.c", "bef"), "a\\.c")
Error in loadNamespace(name) : there is no package called ‘htmlwidgets’
Useful functions for accessing files in packages
- system.file()
- path.package() and normalizePath().
> system.file(package = "batr")
[1] "f:/batr"
> system.file("extdata", package = "batr")
> path.package("batr")
[1] "f:\\batr"
# sometimes it returns the forward slash format for some reason; C:/Program Files/R/R-3.4.0/library/batr
# so it is best to add normalizePath().
> normalizePath(path.package("batr"))
Create R package with devtools and roxygen2
A useful post by Jacob Montgomery. Watch the youtube video there.
The process requires 3 components: RStudio software, devtools and roxygen2 (creating documentation from R code) packages.
MAKING PACKAGES IN R USING DEVTOOLS
R code workflow from Hadley Wickham.
RStudio:addins part 2 - roxygen documentation formatting made easy
devtools cheatsheet (2 pages)
How to use devtools::load_all("FolderName"). load_all() loads any modified R files, and recompile and reload any modified C or Fortran files.
# Step 1
library(devtools)
# Step 2
dir.create(file.path("MyCode", "R"), recursive = TRUE)
cat("foo=function(x){x*2}", file = file.path("MyCode", "R", "foo.R"))
write.dcf(list(Package = "MyCode", Title = "My Code for this project", Description = "To tackle this problem",
Version = "0.0", License = "For my eyes only", Author = "First Last <[email protected]>",
Maintainer = "First Last <[email protected]>"), file = file.path("MyCode", "DESCRIPTION"))
# OR
# create("path/to/package/pkgname")
# create() will create R/ directory, DESCRIPTION and NAMESPACE files.
# Step 3 (C/Fortran code, optional)
dir.create(file.path("MyCode", "src"))
cat("void cfoo(double *a, double *b, double *c){*c=*a+*b;}\n", file = file.path("MyCode",
"src", "cfoo.c"))
cat("useDynLib(MyCode)\n", file = file.path("MyCode", "NAMESPACE"))
# Step 4
load_all("MyCode")
# Step 5
# Modify R/C/Fortran code and run load_all("MyCode")
# Step 6 (Automatically generate the documentation, optional)
document()
# Step 7 (Deployment, optional)
build("MyCode")
# Step 8 (Install the package, optional)
install()
Note:
- load_all("FolderName") will make the FolderName to become like a package to be loaded into the current R session so the 2nd item returned from search() will be "package:FolderName". However, the FolderName does not exist under Program Files/R/R-X.Y.Z/library nor Documents/R/win-library/X.Y/ (Windows OS).
- build("FolderName") will create a tarball in the current directory. User can install the new package for example using Packages -> Install packages from local files on Windows OS.
- For the simplest R package, the source code only contains a file <DESCRIPTION> and a folder <R> with individual R files in the text format.
Binary packages
- No .R files in the R/ directory. There are 3 files that store the parsed functions in an efficient file format. This is the result of loading all the R code and then saving the functions with save().
- A Meta/ directory contains a number of Rds files. These files contain cached metadata about the package, like what topics the help files cover and parsed version of the DESCRIPTION file.
- An html/ directory.
- libs/ directory if you have any code in the src/' directory
- The contents of inst/ are moved to the top-level directory.
What is a library?
A library is simply a directory containing installed packages.
You can use .libPaths() to see which libraries are currently active.
.libPaths() lapply(.libPaths(), dir)
Object names
- Variable and function names should be lower case.
- Use an underscore (_) to separate words within a name (reserve . for S3 methods).
- Camel case is a legitimate alternative, but be consistent! For example, preProcess(), twoClassData, createDataPartition(), trainingRows, trainPredictors, testPredictors, trainClasses, testClasses have been used in Applied Predictive Modeling by Kuhn & Johnson.
- Generally, variable names should be nouns and function names should be verb.
Spacing
- Add a space around the operators +, -, \ and *.
- Include a space around the assignment operators, <- and =.
- Add a space around any comparison operators such as == and <.
Indentation
- Use two spaces to indent code.
- Never mix tabs and spaces.
- RStudio can automatically convert the tab character to spaces (see Tools -> Global options -> Code).
formatR package
Use formatR package to clean up poorly formatted code
install.packages("formatR")
formatR::tidy_dir("R")
Another way is to use the linter package.
install.packages("lintr")
lintr:::lin_package()
Minimal R package for submission
https://stat.ethz.ch/pipermail/r-devel/2013-August/067257.html and CRAN Repository Policy.
Continuous Integration: Travis-CI (Linux, Mac)
- A Beginner's Guide to Travis-CI
- testhat package
- http://johnmuschelli.com/neuroc/getting_ready_for_submission/index.html#61_travis
Continuous Integration: Appveyor (Windows)
- Appveyor is a continuous integration service that builds projects on Windows machines.
- http://johnmuschelli.com/neuroc/getting_ready_for_submission/index.html#62_appveyor
Submit packages to cran
- http://f.briatte.org/r/submitting-packages-to-cran
- https://rmhogervorst.github.io/cleancode/blog/2016/07/09/submtting-to-cran-first-experience.html
- Preparing Your Package for for Submission
- https://builder.r-hub.io/
Build R package faster using multicore
http://www.rexamine.com/2015/07/speeding-up-r-package-installation-process/
The idea is edit the /lib64/R/etc/Renviron file (where /lib64/R/etc/ is the result to a call to the R.home() function in R) and set:
MAKE='make -j 8' # submit 8 jobs at once
Then build R package as regular, for example,
$ time R CMD INSTALL ~/R/stringi --preclean --configure-args='--disable-pkg-config'
suppressPackageStartupMessages
suppressPackageStartupMessages(library("dplyr"))
Tricks
Getting help
- http://stackoverflow.com/questions/tagged/r and R page contains resources.
- https://stat.ethz.ch/pipermail/r-help/
- https://stat.ethz.ch/pipermail/r-devel/
Better Coder/coding, best practices
- http://www.mango-solutions.com/wp/2015/10/10-top-tips-for-becoming-a-better-coder/
- Writing Good R Code and Writing Well
- R Code – Best practices
E-notation
6.022E23 (or 6.022e23) is equivalent to 6.022×10^23
Change default R repository
Edit global Rprofile file. On *NIX platforms, it's located in /usr/lib/R/library/base/R/Rprofile although local .Rprofile settings take precedence.
For example, I can specify the R mirror I like by creating a single line <.Rprofile> file under my home directory.
local({
r = getOption("repos")
r["CRAN"] = "https://cran.rstudio.com/"
options(repos = r)
})
options(continue = " ")
message("Hi MC, loading ~/.Rprofile")
if (interactive()) {
.Last <- function() try(savehistory("~/.Rhistory"))
}
Change the default web browser
When I run help.start() function in LXLE, it cannot find its default web browser (seamonkey).
> help.start()
If the browser launched by 'xdg-open' is already running, it is *not*
restarted, and you must switch to its window.
Otherwise, be patient ...
> /usr/bin/xdg-open: 461: /usr/bin/xdg-open: x-www-browser: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: firefox: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: mozilla: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: epiphany: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: konqueror: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: chromium-browser: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: google-chrome: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: links2: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: links: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: lynx: not found
/usr/bin/xdg-open: 461: /usr/bin/xdg-open: w3m: not found
xdg-open: no method available for opening 'http://127.0.0.1:27919/doc/html/index.html'
The solution is to put
options(browser='seamonkey')
in the .Rprofile of your home directory. If the browser is not in the global PATH, we need to put the full path above.
For one-time only purpose, we can use the browser option in help.start() function:
> help.start(browser="seamonkey")
If the browser launched by 'seamonkey' is already running, it is *not*
restarted, and you must switch to its window.
Otherwise, be patient ...
We can work made a change (or create the file) ~/.Renviron or etc/Renviron. See
- Changing default browser in options().
- https://stat.ethz.ch/R-manual/R-devel/library/utils/html/browseURL.html
Rconsole, Rprofile.site, Renviron.site files
- https://cran.r-project.org/doc/manuals/r-release/R-admin.html (Rprofile.site)
- https://cran.r-project.org/doc/manuals/r-release/R-intro.html (Rprofile.site, Renviron.site, Rconsole (Windows only))
- https://cran.r-project.org/doc/manuals/r-release/R-exts.html (Renviron.site)
- How to store and use webservice keys and authentication details
- Use your .Rprofile to give you important notifications
If we like to install R packages to a personal directory, follow this. Just add the line
R_LIBS_SITE=F:/R/library
to the file R_HOME/etc/x64/Renviron.site.
Note that on Windows OS, R/etc contains
$ ls -l /c/Progra~1/r/r-3.2.0/etc total 142 -rw-r--r-- 1 Administ 1043 Jun 20 2013 Rcmd_environ -rw-r--r-- 1 Administ 1924 Mar 17 2010 Rconsole -rw-r--r-- 1 Administ 943 Oct 3 2011 Rdevga -rw-r--r-- 1 Administ 589 May 20 2013 Rprofile.site -rw-r--r-- 1 Administ 251894 Jan 17 2015 curl-ca-bundle.crt drwxr-xr-x 1 Administ 0 Jun 8 10:30 i386 -rw-r--r-- 1 Administ 1160 Dec 31 2014 repositories -rw-r--r-- 1 Administ 30188 Mar 17 2010 rgb.txt drwxr-xr-x 3 Administ 0 Jun 8 10:30 x64 $ ls /c/Progra~1/r/r-3.2.0/etc/i386 Makeconf $ cat /c/Progra~1/r/r-3.2.0/etc/Rconsole # Optional parameters for the console and the pager # The system-wide copy is in R_HOME/etc. # A user copy can be installed in `R_USER'. ## Style # This can be `yes' (for MDI) or `no' (for SDI). MDI = yes # MDI = no # the next two are only relevant for MDI toolbar = yes statusbar = no ## Font. # Please use only fixed width font. # If font=FixedFont the system fixed font is used; in this case # points and style are ignored. If font begins with "TT ", only # True Type fonts are searched for. font = TT Courier New points = 10 style = normal # Style can be normal, bold, italic # Dimensions (in characters) of the console. rows = 25 columns = 80 # Dimensions (in characters) of the internal pager. pgrows = 25 pgcolumns = 80 # should options(width=) be set to the console width? setwidthonresize = yes # memory limits for the console scrolling buffer, in chars and lines # NB: bufbytes is in bytes for R < 2.7.0, chars thereafter. bufbytes = 250000 buflines = 8000 # Initial position of the console (pixels, relative to the workspace for MDI) # xconsole = 0 # yconsole = 0 # Dimension of MDI frame in pixels # Format (w*h+xorg+yorg) or use -ve w and h for offsets from right bottom # This will come up maximized if w==0 # MDIsize = 0*0+0+0 # MDIsize = 1000*800+100+0 # MDIsize = -50*-50+50+50 # 50 pixels space all round # The internal pager can displays help in a single window # or in multiple windows (one for each topic) # pagerstyle can be set to `singlewindow' or `multiplewindows' pagerstyle = multiplewindows ## Colours for console and pager(s) # (see rwxxxx/etc/rgb.txt for the known colours). background = White normaltext = NavyBlue usertext = Red highlight = DarkRed ## Initial position of the graphics window ## (pixels, <0 values from opposite edge) xgraphics = -25 ygraphics = 0 ## Language for messages language = ## Default setting for console buffering: 'yes' or 'no' buffered = yes
and on Linux
brb@brb-T3500:~$ whereis R R: /usr/bin/R /etc/R /usr/lib/R /usr/bin/X11/R /usr/local/lib/R /usr/share/R /usr/share/man/man1/R.1.gz brb@brb-T3500:~$ ls /usr/lib/R bin COPYING etc lib library modules site-library SVN-REVISION brb@brb-T3500:~$ ls /usr/lib/R/etc javaconf ldpaths Makeconf Renviron Renviron.orig Renviron.site Renviron.ucf repositories Rprofile.site brb@brb-T3500:~$ ls /usr/local/lib/R site-library
and
brb@brb-T3500:~$ cat /usr/lib/R/etc/Rprofile.site
## Emacs please make this -*- R -*-
## empty Rprofile.site for R on Debian
##
## Copyright (C) 2008 Dirk Eddelbuettel and GPL'ed
##
## see help(Startup) for documentation on ~/.Rprofile and Rprofile.site
# ## Example of .Rprofile
# options(width=65, digits=5)
# options(show.signif.stars=FALSE)
# setHook(packageEvent("grDevices", "onLoad"),
# function(...) grDevices::ps.options(horizontal=FALSE))
# set.seed(1234)
# .First <- function() cat("\n Welcome to R!\n\n")
# .Last <- function() cat("\n Goodbye!\n\n")
# ## Example of Rprofile.site
# local({
# # add MASS to the default packages, set a CRAN mirror
# old <- getOption("defaultPackages"); r <- getOption("repos")
# r["CRAN"] <- "http://my.local.cran"
# options(defaultPackages = c(old, "MASS"), repos = r)
#})
brb@brb-T3500:~$ cat /usr/lib/R/etc/Renviron.site
## Emacs please make this -*- R -*-
## empty Renviron.site for R on Debian
##
## Copyright (C) 2008 Dirk Eddelbuettel and GPL'ed
##
## see help(Startup) for documentation on ~/.Renviron and Renviron.site
# ## Example ~/.Renviron on Unix
# R_LIBS=~/R/library
# PAGER=/usr/local/bin/less
# ## Example .Renviron on Windows
# R_LIBS=C:/R/library
# MY_TCLTK="c:/Program Files/Tcl/bin"
# ## Example of setting R_DEFAULT_PACKAGES (from R CMD check)
# R_DEFAULT_PACKAGES='utils,grDevices,graphics,stats'
# # this loads the packages in the order given, so they appear on
# # the search path in reverse order.
brb@brb-T3500:~$
What is the best place to save Rconsole on Windows platform
Put/create the file <Rconsole> under C:/Users/USERNAME/Documents folder so no matter how R was upgraded/downgraded, it always find my preference.
My preferred settings:
- Font: Consolas (it will be shown as "TT Consolas" in Rconsole)
- Size: 12
- background: black
- normaltext: white
- usertext: GreenYellow or orange (close to RStudio's Cobalt theme) or sienna1 or SpringGreen or tan1 or yellow
and others (default options)
- pagebg: white
- pagetext: navy
- highlight: DarkRed
- dataeditbg: white
- dataedittext: navy (View() function)
- dataedituser: red
- editorbg: white (edit() function)
- editortext: black
Saving and loading history automatically: .Rprofile & local()
- http://stat.ethz.ch/R-manual/R-patched/library/utils/html/savehistory.html
- .Rprofile will automatically be loaded when R has started from that directory
- .Rprofile has been created/used by the packrat package to restore a packrat environment. See the packrat/init.R file.
- Customizing Startup from R in Action, Fun with .Rprofile and customizing R startup
- You can also place a .Rprofile file in any directory that you are going to run R from or in the user home directory.
- At startup, R will source the Rprofile.site file. It will then look for a .Rprofile file to source in the current working directory. If it doesn't find it, it will look for one in the user's home directory.
options(continue=" ") # default is "+ "
options(editor="nano") # default is "vi" on Linux
# options(htmlhelp=TRUE)
local((r <- getOption("repos")
r["CRAN"] <- "http://cran.rstudio.com"
options(repos = r)))
.First <- function(){
# library(Hmisc)
cat("\nWelcome at", date(), "\n")
}
.Last <- function(){
cat("\nGoodbye at ", date(), "\n")
}
- https://stackoverflow.com/questions/16734937/saving-and-loading-history-automatically
- The history file will always be read from the $HOME directory and the history file will be overwritten by a new session. These two problems can be solved if we define R_HISTFILE system variable.
- local() function can be used in .Rprofile file to set up the environment even no new variables will be created (change repository, install packages, load libraries, source R files, run system() function, file/directory I/O, etc)
Linux or Mac
In ~/.profile or ~/.bashrc I put:
export R_HISTFILE=~/.Rhistory
In ~/.Rprofile I put:
if (interactive()) {
if (.Platform$OS.type == "unix") .First <- function() try(utils::loadhistory("~/.Rhistory"))
.Last <- function() try(savehistory(file.path(Sys.getenv("HOME"), ".Rhistory")))
}
Windows
If you launch R by clicking its icon from Windows Desktop, the R starts in C:\User\$USER\Documents directory. So we can create a new file .Rprofile in this directory.
if (interactive()) {
.Last <- function() try(savehistory(file.path(Sys.getenv("HOME"), ".Rhistory")))
}
R release versions
rversions: Query the main 'R' 'SVN' repository to find the released versions & dates.
Detect number of running R instances in Windows
- http://stackoverflow.com/questions/15935931/detect-number-of-running-r-instances-in-windows-within-r
C:\Program Files\R>tasklist /FI "IMAGENAME eq Rscript.exe" INFO: No tasks are running which match the specified criteria. C:\Program Files\R>tasklist /FI "IMAGENAME eq Rgui.exe" Image Name PID Session Name Session# Mem Usage ========================= ======== ================ =========== ============ Rgui.exe 1096 Console 1 44,712 K C:\Program Files\R>tasklist /FI "IMAGENAME eq Rserve.exe" Image Name PID Session Name Session# Mem Usage ========================= ======== ================ =========== ============ Rserve.exe 6108 Console 1 381,796 K
In R, we can use
> system('tasklist /FI "IMAGENAME eq Rgui.exe" ', intern = TRUE)
[1] ""
[2] "Image Name PID Session Name Session# Mem Usage"
[3] "========================= ======== ================ =========== ============"
[4] "Rgui.exe 1096 Console 1 44,804 K"
> length(system('tasklist /FI "IMAGENAME eq Rgui.exe" ', intern = TRUE))-3
Editor
http://en.wikipedia.org/wiki/R_(programming_language)#Editors_and_IDEs
- Emacs + ESS. The ESS is useful in the case I want to tidy R code (the tidy_source() function in the formatR package sometimes gives errors; eg when I tested it on an R file like <GetComparisonResults.R> from BRB-ArrayTools v4.4 stable).
- Rstudio - editor/R terminal/R graphics/file browser/package manager. The new version (0.98) also provides a new feature for debugging step-by-step. See also RStudio Tricks
- geany - I like the feature that it shows defined functions on the side panel even for R code. RStudio can also do this (see the bottom of the code panel).
- Rgedit which includes a feature of splitting screen into two panes and run R in the bottom panel. See here.
- Komodo IDE with browser preview http://www.youtube.com/watch?v=wv89OOw9roI at 4:06 and http://docs.activestate.com/komodo/4.4/editor.html
GUI for Data Analysis
Rcmdr
http://cran.r-project.org/web/packages/Rcmdr/index.html
Deducer
http://cran.r-project.org/web/packages/Deducer/index.html
Scope
See
- Assignments within functions in the An Introduction to R manual.
- source() does not work like C's preprocessor where statements in header files will be literally inserted into the code. It does not work when you define a variable in a function but want to use it outside the function (even through source())
## foo.R ##
cat(ArrayTools, "\n")
## End of foo.R
# 1. Error
predict <- function() {
ArrayTools <- "C:/Program Files" # or through load() function
source("foo.R") # or through a function call; foo()
}
predict() # Object ArrayTools not found
# 2. OK. Make the variable global
predict <- function() {
ArrayTools <<- "C:/Program Files'
source("foo.R")
}
predict()
ArrayTools
# 3. OK. Create a global variable
ArrayTools <- "C:/Program Files"
predict <- function() {
source("foo.R")
}
predict()
Note that any ordinary assignments done within the function are local and temporary and are lost after exit from the function.
Example 1.
> ttt <- data.frame(type=letters[1:5], JpnTest=rep("999", 5), stringsAsFactors = F)
> ttt
type JpnTest
1 a 999
2 b 999
3 c 999
4 d 999
5 e 999
> jpntest <- function() { ttt$JpnTest[1] ="N5"; print(ttt)}
> jpntest()
type JpnTest
1 a N5
2 b 999
3 c 999
4 d 999
5 e 999
> ttt
type JpnTest
1 a 999
2 b 999
3 c 999
4 d 999
5 e 999
Example 2. How can we set global variables inside a function? The answer is to use the "<<-" operator or assign(, , envir = .GlobalEnv) function.
Other resource: Advanced R by Hadley Wickham.
Example 3. Writing functions in R, keeping scoping in mind
New environment
http://adv-r.had.co.nz/Environments.html
Run the same function on a bunch of R objects
mye = new.env() load(<filename>, mye) for(n in names(mye)) n = as_tibble(mye[[n]])
Speedup R code
- Strategies to speedup R code from DataScience+
Profiler
(Video) Understand Code Performance with the profiler
&& vs &
See https://www.rdocumentation.org/packages/base/versions/3.5.1/topics/Logic.
The shorter form performs elementwise comparisons in much the same way as arithmetic operators. The longer form evaluates left to right examining only the first element of each vector.
Vectorization
- https://en.wikipedia.org/wiki/Vectorization_%28mathematics%29
- http://www.noamross.net/blog/2014/4/16/vectorization-in-r--why.html
- https://github.com/vsbuffalo/devnotes/wiki/R-and-Vectorization
split()
split() can be used to split columns or rows. See How to split a data frame?
- Split rows of a data frame/matrix
split(mtcars,mtcars$cyl)
- Split columns of a data frame/matrix.
- Example 1.
ma <- cbind(x = 1:10, y = (-4:5)^2, z = 11:20) split(ma, cbind(rep(1,10), rep(2, 10), rep(1,10))) # $`1` # [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 # # $`2` # [1] 16 9 4 1 0 1 4 9 16 25
- Example 2. sapply() + split() to merge columns. See below 'Mean of duplicated columns' for more detail.
- Example 1.
Mean of duplicated columns
- Reduce columns of a matrix by a function in R
x <- matrix(1:60, nr=10); x colnames(x) <- c("A","A", "b", "b", "b", "c") res <- sapply(split(1:ncol(x), colnames(x)), function(i) rowSums(x[, i, drop = F])) res # vapply() is safter than sapply(). # The 3rd arg in vapply() is a template of the return value. res2 <- vapply(split(1:ncol(x), colnames(x)), function(i) rowSums(x[, i, drop = F]), rep(0, nrow(x))) - rowsum() on a transposed matrix.
Mean of duplicated rows
- rowsum()
- use ave() and unique()
- data.table package
- plyr package
- aggregate() function. Too slow! http://slowkow.com/2015/01/28/data-table-aggregate/. Don't use aggregate post.
> attach(mtcars) dim(mtcars) [1] 32 11 > head(mtcars) mpg cyl disp hp drat wt qsec vs am gear carb Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4 Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4 Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1 Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1 Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2 Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1 > aggdata <-aggregate(mtcars, by=list(cyl,vs), FUN=mean, na.rm=TRUE) > print(aggdata) Group.1 Group.2 mpg cyl disp hp drat wt qsec vs 1 4 0 26.00000 4 120.30 91.0000 4.430000 2.140000 16.70000 0 2 6 0 20.56667 6 155.00 131.6667 3.806667 2.755000 16.32667 0 3 8 0 15.10000 8 353.10 209.2143 3.229286 3.999214 16.77214 0 4 4 1 26.73000 4 103.62 81.8000 4.035000 2.300300 19.38100 1 5 6 1 19.12500 6 204.55 115.2500 3.420000 3.388750 19.21500 1 am gear carb 1 1.0000000 5.000000 2.000000 2 1.0000000 4.333333 4.666667 3 0.1428571 3.285714 3.500000 4 0.7000000 4.000000 1.500000 5 0.0000000 3.500000 2.500000 > detach(mtcars) # Another example: select rows with a minimum value from a certain column (yval in this case) > mydf <- read.table(header=T, text=' id xval yval A 1 1 A -2 2 B 3 3 B 4 4 C 5 5 ') > x = mydf$xval > y = mydf$yval > aggregate(mydf[, c(2,3)], by=list(id=mydf$id), FUN=function(x) x[which.min(y)]) id xval yval 1 A 1 1 2 B 3 3 3 C 5 5
Apply family
Vectorize, aggregate, apply, by, eapply, lapply, mapply, rapply, replicate, scale, sapply, split, tapply, and vapply. Check out this.
The following list gives a hierarchical relationship among these functions.
- apply(X, MARGIN, FUN, ...) – Apply a Functions Over Array Margins
- tapply(X, INDEX, FUN = NULL, ..., default = NA, simplify = TRUE) – Apply a Function Over a "Ragged" Array
- by(data, INDICES, FUN, ..., simplify = TRUE) - Apply a Function to a Data Frame Split by Factors
- aggregate(x, by, FUN, ..., simplify = TRUE, drop = TRUE) - Compute Summary Statistics of Data Subsets
- lapply(X, FUN, ...) – Apply a Function over a List or Vector
- sapply(X, FUN, ..., simplify = TRUE, USE.NAMES = TRUE) – Apply a Function over a List or Vector
- replicate(n, expr, simplify = "array")
- mapply(FUN, ..., MoreArgs = NULL, SIMPLIFY = TRUE, USE.NAMES = TRUE) – Multivariate version of sapply
- Vectorize(FUN, vectorize.args = arg.names, SIMPLIFY = TRUE, USE.NAMES = TRUE) - Vectorize a Scalar Function
- vapply(X, FUN, FUN.VALUE, ..., USE.NAMES = TRUE) – similar to sapply, but has a pre-specified type of return value
- sapply(X, FUN, ..., simplify = TRUE, USE.NAMES = TRUE) – Apply a Function over a List or Vector
- rapply(object, f, classes = "ANY", deflt = NULL, how = c("unlist", "replace", "list"), ...) – A recursive version of lapply
- eapply(env, FUN, ..., all.names = FALSE, USE.NAMES = TRUE) – Apply a Function over values in an environment
Note that, apply's performance is not always better than a for loop. See
- http://tolstoy.newcastle.edu.au/R/help/06/05/27255.html (answered by Brian Ripley)
- https://stat.ethz.ch/pipermail/r-help/2014-October/422455.html (has one example)
The package 'pbapply' creates a text-mode progress bar - it works on any platforms. On Windows platform, check out this post. It uses winProgressBar() and setWinProgressBar() functions.
Progress bar
What is the cost of a progress bar in R?
lapply and its friends Map(), Reduce(), Filter() from the base package for manipulating lists
- Examples of using lapply() + split() on a data frame. See rollingyours.wordpress.com.
- mapply() documentation. Use mapply() to merge lists.
- Map() and Reduce() in functional programming
- Map(), Reduce(), and Filter() from Advanced R by Hadley
- If you have two or more lists (or data frames) that you need to process in parallel, use Map(). One good example is to compute the weighted.mean() function that requires two input objects. Map() is similar to mapply() function and is more concise than lapply(). Advanced R has a comment that Map() is better than mapply().
# Syntax: Map(f, ...) xs <- replicate(5, runif(10), simplify = FALSE) ws <- replicate(5, rpois(10, 5) + 1, simplify = FALSE) Map(weighted.mean, xs, ws) # instead of a more clumsy way lapply(seq_along(xs), function(i) { weighted.mean(xs[[i]], ws[[i]]) }) - Reduce() reduces a vector, x, to a single value by recursively calling a function, f, two arguments at a time. A good example of using Reduce() function is to read a list of matrix files and merge them. See How to combine multiple matrix frames into one using R?
# Syntax: Reduce(f, x, ...) > m1 <- data.frame(id=letters[1:4], val=1:4) > m2 <- data.frame(id=letters[2:6], val=2:6) > merge(m1, m2, "id", all = T) id val.x val.y 1 a 1 NA 2 b 2 2 3 c 3 3 4 d 4 4 5 e NA 5 6 f NA 6 > m <- list(m1, m2) > Reduce(function(x,y) merge(x,y, "id",all=T), m) id val.x val.y 1 a 1 NA 2 b 2 2 3 c 3 3 4 d 4 4 5 e NA 5 6 f NA 6
- If you have two or more lists (or data frames) that you need to process in parallel, use Map(). One good example is to compute the weighted.mean() function that requires two input objects. Map() is similar to mapply() function and is more concise than lapply(). Advanced R has a comment that Map() is better than mapply().
- Playing Map() and Reduce() in R – Subsetting - using parallel and future packages. Union Multiple Data.Frames with Different Column Names
sapply & vapply
- This discusses why vapply is safer and faster than sapply.
- Vector output: sapply and vapply from Advanced R (Hadley Wickham).
- THOSE “OTHER” APPLY FUNCTIONS…. rapply(), vapply() and eapply() are covered.
rapply - recursive version of lapply
- http://4dpiecharts.com/tag/recursive/
- Search in R source code. Mainly r-source/src/library/stats/R/dendrogram.R.
replicate
https://www.datacamp.com/community/tutorials/tutorial-on-loops-in-r
> replicate(5, rnorm(3))
[,1] [,2] [,3] [,4] [,5]
[1,] 0.2509130 -0.3526600 -0.3170790 1.064816 -0.53708856
[2,] 0.5222548 1.5343319 0.6120194 -1.811913 -1.09352459
[3,] -1.9905533 -0.8902026 -0.5489822 1.308273 0.08773477
See parSapply() for a parallel version of replicate().
Vectorize
> rweibull(1, 1, c(1, 2)) # no error but not sure what it gives?
[1] 2.17123
> Vectorize("rweibull")(n=1, shape = 1, scale = c(1, 2))
[1] 1.6491761 0.9610109
https://blogs.msdn.microsoft.com/gpalem/2013/03/28/make-vectorize-your-friend-in-r/
myfunc <- function(a, b) a*b
myfunc(1, 2) # 2
myfunc(3, 5) # 15
myfunc(c(1,3), c(2,5)) # 2 15
Vectorize(myfunc)(c(1,3), c(2,5)) # 2 15
myfunc2 <- function(a, b) if (length(a) == 1) a * b else NA
myfunc2(1, 2) # 2
myfunc2(3, 5) # 15
myfunc2(c(1,3), c(2,5)) # NA
Vectorize(myfunc2)(c(1, 3), c(2, 5)) # 2 15
Vectorize(myfunc2)(c(1, 3, 6), c(2, 5)) # 2 15 12
# parameter will be re-used
plyr and dplyr packages
Practical Data Science for Stats - a PeerJ Collection
The Split-Apply-Combine Strategy for Data Analysis (plyr package) in J. Stat Software.
A quick introduction to plyr with a summary of apply functions in R and compare them with functions in plyr package.
- plyr has a common syntax -- easier to remember
- plyr requires less code since it takes care of the input and output format
- plyr can easily be run in parallel -- faster
Tutorials
- Introduction to dplyr from http://dplyr.tidyverse.org/.
- A video of dplyr package can be found on vimeo.
- Hands-on dplyr tutorial for faster data manipulation in R from dataschool.io.
Examples of using dplyr:
- Medicines under evaluation
- CBI GEO Metadata Survey
- Logistic Regression by Page Piccinini. mutate(), inner_join() and %>%.
- DESeq2 post analysis select(), gather(), arrange() and %>%.
tibble
Tibbles are data frames, but slightly tweaked to work better in the tidyverse.
> data(pew, package = "efficient")
> dim(pew)
[1] 18 10
> class(pew) # tibble is also a data frame!!
[1] "tbl_df" "tbl" "data.frame"
> tidyr::gather(pew, key=Income, value = Count, -religion) # make wide tables long
# A tibble: 162 x 3
religion Income Count
<chr> <chr> <int>
1 Agnostic <$10k 27
2 Atheist <$10k 12
...
> mean(tidyr::gather(pew, key=Income, value = Count, -religion)[, 3])
[1] NA
Warning message:
In mean.default(tidyr::gather(pew, key = Income, value = Count, :
argument is not numeric or logical: returning NA
> mean(tidyr::gather(pew, key=Income, value = Count, -religion)[[3]])
[1] 181.6975
If we try to do a match on some column of a tibble object, we will get zero matches. The issue is we cannot use an index to get a tibble column.
Subsetting: to extract a column from a tibble object, use [[ or $ or dplyr::pull().
TibbleObject$VarName # OR TibbleObject[["VarName"]] # OR pull(TibbleObject, VarName) # won't be a tibble object anymore
llply()
llply is equivalent to lapply except that it will preserve labels and can display a progress bar. This is handy if we want to do a crazy thing.
LLID2GOIDs <- lapply(rLLID, function(x) get("org.Hs.egGO")[[x]])
where rLLID is a list of entrez ID. For example,
get("org.Hs.egGO")[["6772"]]
returns a list of 49 GOs.
ddply()
http://lamages.blogspot.com/2012/06/transforming-subsets-of-data-in-r-with.html
ldply()
An R Script to Automatically download PubMed Citation Counts By Year of Publication
Using R's set.seed() to set seeds for use in C/C++ (including Rcpp)
http://rorynolan.rbind.io/2018/09/30/rcsetseed/
get_seed()
See the same blog
get_seed <- function() {
sample.int(.Machine$integer.max, 1)
}
set.seed(), for loop and saving random seeds
http://r.789695.n4.nabble.com/set-seed-and-for-loop-td3585857.html. This question is legitimate when we want to debug on a certain iteration.
set.seed(1001)
data <- vector("list", 30)
seeds <- vector("list", 30)
for(i in 1:30) {
seeds[[i]] <- .Random.seed
data[[i]] <- runif(5)
}
.Random.seed <- seeds[[23]] # restore
data.23 <- runif(5)
data.23
data[[23]]
- Duncan Murdoch: This works in this example, but wouldn't work with all RNGs, because some of them save state outside of .Random.seed. See ?.Random.seed for details.
- Uwe Ligges's comment: set.seed() actually generates a seed. See ?set.seed that points us to .Random.seed (and relevant references!) which contains the actual current seed.
- Petr Savicky's comment is also useful in the situation when it is not difficult to re-generate the data.
mclapply() and parLapply()
mclapply() from the 'parallel' package is a mult-core version of lapply()
- Be providing the number of cores in mclapply() using mc.cores argument (2 is used by default)
- Be careful on the need and the side-effect of using "L'Ecuyer-CMRG" seed.
- R doesn't reset the seed when “L'Ecuyer-CMRG” RNG is used?
library(parallel) system.time(mclapply(1:1e4L, function(x) rnorm(x))) system.time(mclapply(1:1e4L, function(x) rnorm(x), mc.cores = 4)) set.seed(1234) mclapply(1:3, function(x) rnorm(x)) set.seed(1234) mclapply(1:3, function(x) rnorm(x)) # cannot reproduce the result set.seed(123, "L'Ecuyer") mclapply(1:3, function(x) rnorm(x)) mclapply(1:3, function(x) rnorm(x)) # results are not changed once we have run set.seed( , "L'Ecuyer") set.seed(1234) # use set.seed() in order to get a new reproducible result mclapply(1:3, function(x) rnorm(x)) mclapply(1:3, function(x) rnorm(x)) # results are not changed
Note
- R doesn't reset the seed when “L'Ecuyer-CMRG” RNG is used?
- Windows OS can not use mclapply(). The mclapply() implementation relies on forking and Windows does not support forking. mclapply from the parallel package is implemented as a serial function on Windows systems. The parallelsugar package was created based on the above idea.
- Another choice for Windows OS is to use parLapply() function in parallel package.
- Understanding the differences between mclapply and parLapply in R You don't have to worry about reproducing your environment on each of the cluster workers if mclapply() is used.
ncores <- as.integer( Sys.getenv('NUMBER_OF_PROCESSORS') ) cl <- makeCluster(getOption("cl.cores", ncores)) LLID2GOIDs2 <- parLapply(cl, rLLID, function(x) { library(org.Hs.eg.db); get("org.Hs.egGO")[[x]]} ) stopCluster(cl)It does work. Cut the computing time from 100 sec to 29 sec on 4 cores.
mclapply() vs foreach()
https://stackoverflow.com/questions/44806048/r-mclapply-vs-foreach
parallel vs doParallel package
parallelsugar package
If we load parallelsugar, the default implementation of parallel::mclapply, which used fork based clusters, will be overwritten by parallelsugar::mclapply, which is implemented with socket clusters.
library(parallel)
system.time( mclapply(1:4, function(xx){ Sys.sleep(10) }) )
## user system elapsed
## 0.00 0.00 40.06
library(parallelsugar)
##
## Attaching package: ‘parallelsugar’
##
## The following object is masked from ‘package:parallel’:
##
## mclapply
system.time( mclapply(1:4, function(xx){ Sys.sleep(10) }) )
## user system elapsed
## 0.04 0.08 12.98
Regular Expression
See here.
Read rrd file
- https://en.wikipedia.org/wiki/RRDtool
- http://oss.oetiker.ch/rrdtool/
- https://github.com/pldimitrov/Rrd
- http://plamendimitrov.net/blog/2014/08/09/r-package-for-working-with-rrd-files/
file, connection
- cat() and scan() (read data into a vector or list from the console or file)
- read() and write()
- read.table() and write.table()
Clipboard (?connections) & textConnection()
source("clipboard")
read.table("clipboard")
- On Windows, we can use readClipboard() and writeClipboard().
- reading/writing clipboard method seems not quite stable on Linux/macOS. So the alternative is to use the textConnection() function:
x <- read.delim(textConnection("<USE_KEYBOARD_TO_PASTE_FROM_CLIPBOARD>"))An example is to copy data from this post. In this case we need to use read.table() instead of read.delim().
read/manipulate binary data
- x <- readBin(fn, raw(), file.info(fn)$size)
- rawToChar(x[1:16])
- See Biostrings C API
String Manipulation
- ebook by Gaston Sanchez.
- A guide to working with character data in R (6/22/2018)
- Chapter 7 of the book 'Data Manipulation with R' by Phil Spector.
- Chapter 7 of the book 'R Cookbook' by Paul Teetor.
- Chapter 2 of the book 'Using R for Data Management, Statistical Analysis and Graphics' by Horton and Kleinman.
- http://www.endmemo.com/program/R/deparse.php. It includes lots of examples for each R function it lists.
HTTPs connection
HTTPS connection becomes default in R 3.2.2. See
- http://blog.rstudio.org/2015/08/17/secure-https-connections-for-r/
- http://blog.revolutionanalytics.com/2015/08/good-advice-for-security-with-r.html
R 3.3.2 patched The internal methods of ‘download.file()’ and ‘url()’ now report if they are unable to follow the redirection of a ‘http://’ URL to a ‘https://’ URL (rather than failing silently)
setInternet2
There was a bug in ftp downloading in R 3.2.2 (r69053) Windows though it is fixed now in R 3.2 patch.
Read the discussion reported on 8/8/2015. The error only happened on ftp not http connection. The final solution is explained in this post. The following demonstrated the original problem.
url <- paste0("ftp://ftp.ncbi.nlm.nih.gov/genomes/ASSEMBLY_REPORTS/All/",
"GCF_000001405.13.assembly.txt")
f1 <- tempfile()
download.file(url, f1)
It seems the bug was fixed in R 3.2-branch. See 8/16/2015 patch r69089 where a new argument INTERNET_FLAG_PASSIVE was added to InternetOpenUrl() function of wininet library. This article and this post explain differences of active and passive FTP.
The following R command will show the exact svn revision for the R you are currently using.
R.Version()$"svn rev"
If setInternet2(T), then https protocol is supported in download.file().
When setInternet(T) is enabled by default, download.file() does not work for ftp protocol (this is used in getGEO() function of the GEOquery package). If I use setInternet(F), download.file() works again for ftp protocol.
The setInternet2() function is defined in R> src> library> utils > R > windows > sysutils.R.
R up to 3.2.2
setInternet2 <- function(use = TRUE) .Internal(useInternet2(use))
See also
- <src/include/Internal.h> (declare do_setInternet2()),
- <src/main/names.c> (show do_setInternet2() in C)
- <src/main/internet.c> (define do_setInternet2() in C).
Note that: setInternet2(T) becomes default in R 3.2.2. To revert to the previous default use setInternet2(FALSE). See the <doc/NEWS.pdf> file. If we use setInternet2(F), then it solves the bug of getGEO() error. But it disables the https file download using the download.file() function. In R < 3.2.2, it is also possible to download from https by setIneternet2(T).
R 3.3.0
setInternet2 <- function(use = TRUE) {
if(!is.na(use)) stop("use != NA is defunct")
NA
}
Note that setInternet2.Rd says As from \R 3.3.0 it changes nothing, and only \code{use = NA} is accepted. Also NEWS.Rd says setInternet2() has no effect and will be removed in due course.
File operation
- list.files()
- file.info()
- dir.create()
- file.create()
- file.copy()
read/download/source a file from internet
Simple text file http
retail <- read.csv("http://robjhyndman.com/data/ausretail.csv",header=FALSE)
Zip file and url() function
con = gzcon(url('http://www.systematicportfolio.com/sit.gz', 'rb'))
source(con)
close(con)
Here url() function is like file(), gzfile(), bzfile(), xzfile(), unz(), pipe(), fifo(), socketConnection(). They are used to create connections. By default, the connection is not opened (except for ‘socketConnection’), but may be opened by setting a non-empty value of argument ‘open’. See ?url.
Another example of using url() is
load(url("http:/www.example.com/example.RData"))
downloader package
This package provides a wrapper for the download.file function, making it possible to download files over https on Windows, Mac OS X, and other Unix-like platforms. The RCurl package provides this functionality (and much more) but can be difficult to install because it must be compiled with external dependencies. This package has no external dependencies, so it is much easier to install.
Google drive file based on https using RCurl package
require(RCurl)
myCsv <- getURL("https://docs.google.com/spreadsheet/pub?hl=en_US&hl=en_US&key=0AkuuKBh0jM2TdGppUFFxcEdoUklCQlJhM2kweGpoUUE&single=true&gid=0&output=csv")
read.csv(textConnection(myCsv))
Google sheet file using googlesheets package
Reading data from google sheets into R
Github files https using RCurl package
- http://support.rstudio.org/help/kb/faq/configuring-r-to-use-an-http-proxy
- http://tonybreyal.wordpress.com/2011/11/24/source_https-sourcing-an-r-script-from-github/
x = getURL("https://gist.github.com/arraytools/6671098/raw/c4cb0ca6fe78054da8dbe253a05f7046270d5693/GeneIDs.txt",
ssl.verifypeer = FALSE)
read.table(text=x)
- gistr package
Create publication tables using tables package
See p13 for example in http://www.ianwatson.com.au/stata/tabout_tutorial.pdf
R's tables packages is the best solution. For example,
> library(tables)
> tabular( (Species + 1) ~ (n=1) + Format(digits=2)*
+ (Sepal.Length + Sepal.Width)*(mean + sd), data=iris )
Sepal.Length Sepal.Width
Species n mean sd mean sd
setosa 50 5.01 0.35 3.43 0.38
versicolor 50 5.94 0.52 2.77 0.31
virginica 50 6.59 0.64 2.97 0.32
All 150 5.84 0.83 3.06 0.44
> str(iris)
'data.frame': 150 obs. of 5 variables:
$ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
$ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
$ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
$ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
$ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
and
# This example shows some of the less common options
> Sex <- factor(sample(c("Male", "Female"), 100, rep=TRUE))
> Status <- factor(sample(c("low", "medium", "high"), 100, rep=TRUE))
> z <- rnorm(100)+5
> fmt <- function(x) {
s <- format(x, digits=2)
even <- ((1:length(s)) %% 2) == 0
s[even] <- sprintf("(%s)", s[even])
s
}
> tabular( Justify(c)*Heading()*z*Sex*Heading(Statistic)*Format(fmt())*(mean+sd) ~ Status )
Status
Sex Statistic high low medium
Female mean 4.88 4.96 5.17
sd (1.20) (0.82) (1.35)
Male mean 4.45 4.31 5.05
sd (1.01) (0.93) (0.75)
See also a collection of R packages related to reproducible research in http://cran.r-project.org/web/views/ReproducibleResearch.html
Extracting tables from PDFs
- extracting Tables from PDFs in R using Tabulizer. This needs the rJava package. Some issue came out on my macOS 10.12 Sierra. Library not loaded: /Library/Java/JavaVirtualMachines/jdk-9.jdk/Contents/Home/lib/server/libjvm.dylib. Referenced from: /Users/XXXXXXX/Library/R/3.5/library/rJava/libs/rJava.so.
- pdftools - Text Extraction, Rendering and Converting of PDF Documents. pdf_text() and pdf_data() functions.
library(pdftools) pdf_file <- "https://github.com/ropensci/tabulizer/raw/master/inst/examples/data.pdf" txt <- pdf_text(pdf_file) # length = number of pages # Suppose the table we are interested in is on page 1 cat(txt[1]) # Good but not in a data frame format pdf_data(pdf_file)[[1]] # data frame/tibble format
Create flat tables in R console using ftable()
> ftable(Titanic, row.vars = 1:3)
Survived No Yes
Class Sex Age
1st Male Child 0 5
Adult 118 57
Female Child 0 1
Adult 4 140
2nd Male Child 0 11
Adult 154 14
Female Child 0 13
Adult 13 80
3rd Male Child 35 13
Adult 387 75
Female Child 17 14
Adult 89 76
Crew Male Child 0 0
Adult 670 192
Female Child 0 0
Adult 3 20
> ftable(Titanic, row.vars = 1:2, col.vars = "Survived")
Survived No Yes
Class Sex
1st Male 118 62
Female 4 141
2nd Male 154 25
Female 13 93
3rd Male 422 88
Female 106 90
Crew Male 670 192
Female 3 20
> ftable(Titanic, row.vars = 2:1, col.vars = "Survived")
Survived No Yes
Sex Class
Male 1st 118 62
2nd 154 25
3rd 422 88
Crew 670 192
Female 1st 4 141
2nd 13 93
3rd 106 90
Crew 3 20
> str(Titanic)
table [1:4, 1:2, 1:2, 1:2] 0 0 35 0 0 0 17 0 118 154 ...
- attr(*, "dimnames")=List of 4
..$ Class : chr [1:4] "1st" "2nd" "3rd" "Crew"
..$ Sex : chr [1:2] "Male" "Female"
..$ Age : chr [1:2] "Child" "Adult"
..$ Survived: chr [1:2] "No" "Yes"
> x <- ftable(mtcars[c("cyl", "vs", "am", "gear")])
> x
gear 3 4 5
cyl vs am
4 0 0 0 0 0
1 0 0 1
1 0 1 2 0
1 0 6 1
6 0 0 0 0 0
1 0 2 1
1 0 2 2 0
1 0 0 0
8 0 0 12 0 0
1 0 0 2
1 0 0 0 0
1 0 0 0
> ftable(x, row.vars = c(2, 4))
cyl 4 6 8
am 0 1 0 1 0 1
vs gear
0 3 0 0 0 0 12 0
4 0 0 0 2 0 0
5 0 1 0 1 0 2
1 3 1 0 2 0 0 0
4 2 6 2 0 0 0
5 0 1 0 0 0 0
>
> ## Start with expressions, use table()'s "dnn" to change labels
> ftable(mtcars$cyl, mtcars$vs, mtcars$am, mtcars$gear, row.vars = c(2, 4),
dnn = c("Cylinders", "V/S", "Transmission", "Gears"))
Cylinders 4 6 8
Transmission 0 1 0 1 0 1
V/S Gears
0 3 0 0 0 0 12 0
4 0 0 0 2 0 0
5 0 1 0 1 0 2
1 3 1 0 2 0 0 0
4 2 6 2 0 0 0
5 0 1 0 0 0 0
addmargins
Puts Arbitrary Margins On Multidimensional Tables Or Arrays
tracemem, data type, copy
How to avoid copying a long vector
Tell if the current R is running in 32-bit or 64-bit mode
8 * .Machine$sizeof.pointer
where sizeof.pointer returns the number of *bytes* in a C SEXP type and '8' means number of bits per byte.
32- and 64-bit
See R-admin.html.
- For speed you may want to use a 32-bit build, but to handle large datasets a 64-bit build.
- Even on 64-bit builds of R there are limits on the size of R objects, some of which stem from the use of 32-bit integers (especially in FORTRAN code). For example, the dimensionas of an array are limited to 2^31 -1.
- Since R 2.15.0, it is possible to select '64-bit Files' from the standard installer even on a 32-bit version of Windows (2012/3/30).
Handling length 2^31 and more in R 3.0.0
From R News for 3.0.0 release:
There is a subtle change in behaviour for numeric index values 2^31 and larger. These never used to be legitimate and so were treated as NA, sometimes with a warning. They are now legal for long vectors so there is no longer a warning, and x[2^31] <- y will now extend the vector on a 64-bit platform and give an error on a 32-bit one.
In R 2.15.2, if I try to assign a vector of length 2^31, I will get an error
> x <- seq(1, 2^31) Error in from:to : result would be too long a vector
However, for R 3.0.0 (tested on my 64-bit Ubuntu with 16GB RAM. The R was compiled by myself):
> system.time(x <- seq(1,2^31))
user system elapsed
8.604 11.060 120.815
> length(x)
[1] 2147483648
> length(x)/2^20
[1] 2048
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 183823 9.9 407500 21.8 350000 18.7
Vcells 2147764406 16386.2 2368247221 18068.3 2148247383 16389.9
>
Note:
- 2^31 length is about 2 Giga length. It takes about 16 GB (2^31*8/2^20 MB) memory.
- On Windows, it is almost impossible to work with 2^31 length of data if the memory is less than 16 GB because virtual disk on Windows does not work well. For example, when I tested on my 12 GB Windows 7, the whole Windows system freezes for several minutes before I force to power off the machine.
- My slide in http://goo.gl/g7sGX shows the screenshots of running the above command on my Ubuntu and RHEL machines. As you can see the linux is pretty good at handling large (> system RAM) data. That said, as long as your linux system is 64-bit, you can possibly work on large data without too much pain.
- For large dataset, it makes sense to use database or specially crafted packages like bigmemory or ff or bigstatsr.
NA in index
- Question: what is seq(1, 3)[c(1, 2, NA)]?
Answer: It will reserve the element with NA in indexing and return the value NA for it.
- Question: What is TRUE & NA?
Answer: NA
- Question: What is FALSE & NA?
Answer: FALSE
- Question: c("A", "B", NA) != "" ?
Answer: TRUE TRUE NA
- Question: which(c("A", "B", NA) != "") ?
Answer: 1 2
- Question: c(1, 2, NA) != "" & !is.na(c(1, 2, NA)) ?
Answer: TRUE TRUE FALSE
- Question: c("A", "B", NA) != "" & !is.na(c("A", "B", NA)) ?
Answer: TRUE TRUE FALSE
Conclusion: In order to exclude empty or NA for numerical or character data type, we can use which() or a convenience function keep.complete(x) <- function(x) x != "" & !is.na(x). This will guarantee return logical values and not contain NAs.
Don't just use x != "" OR !is.na(x).
Constant
Add 'L' after a constant. For example,
for(i in 1L:n) { }
if (max.lines > 0L) { }
label <- paste0(n-i+1L, ": ")
n <- length(x); if(n == 0L) { }
Data frame
stringsAsFactors = FALSE
http://www.win-vector.com/blog/2018/03/r-tip-use-stringsasfactors-false/
data.frame to vector
> a= matrix(1:6, 2,3)
> rownames(a) <- c("a", "b")
> colnames(a) <- c("x", "y", "z")
> a
x y z
a 1 3 5
b 2 4 6
> unlist(data.frame(a))
x1 x2 y1 y2 z1 z2
1 2 3 4 5 6
merge
How to perform merges (joins) on two or more data frames with base R, tidyverse and data.table
matrix vs data.frame
ip1 <- installed.packages()[,c(1,3:4)] # class(ip1) = 'matrix' unique(ip1$Priority) # Error in ip1$Priority : $ operator is invalid for atomic vectors unique(ip1[, "Priority"]) # OK ip2 <- as.data.frame(installed.packages()[,c(1,3:4)], stringsAsFactors = FALSE) # matrix -> data.frame unique(ip2$Priority) # OK
matrix multiply a vector
> matrix(1:6, 3,2)
[,1] [,2]
[1,] 1 4
[2,] 2 5
[3,] 3 6
> matrix(1:6, 3,2) * c(1,2,3)
[,1] [,2]
[1,] 1 4
[2,] 4 10
[3,] 9 18
> matrix(1:6, 3,2) * c(1,2,3,4)
[,1] [,2]
[1,] 1 16
[2,] 4 5
[3,] 9 12
Print a vector by suppressing names
Use unname.
format.pval
> args(format.pval)
function (pv, digits = max(1L, getOption("digits") - 2L), eps = .Machine$double.eps,
na.form = "NA", ...)
> format.pval(c(stats::runif(5), pi^-100, NA))
[1] "0.19571" "0.46793" "0.71696" "0.93200" "0.74485" "< 2e-16" "NA"
> format.pval(c(0.1, 0.0001, 1e-27))
[1] "1e-01" "1e-04" "<2e-16"
Disable scientific notation in printing
> numer = 29707; denom = 93874 > c(numer/denom, numer, denom) [1] 3.164561e-01 2.970700e+04 9.387400e+04 # Method 1. Without changing the global option > format(c(numer/denom, numer, denom), scientific=FALSE) [1] " 0.3164561" "29707.0000000" "93874.0000000" # Method 2. Change the global option > options(scipen=999) > numer/denom [1] 0.3164561 > c(numer/denom, numer, denom) [1] 0.3164561 29707.0000000 93874.0000000 > c(4/5, numer, denom) [1] 0.8 29707.0 93874.0
sprintf
Format number as fixed width, with leading zeros
- https://stackoverflow.com/questions/8266915/format-number-as-fixed-width-with-leading-zeros
- https://stackoverflow.com/questions/14409084/pad-with-leading-zeros-to-common-width?rq=1
# sprintf()
a <- seq(1,101,25)
sprintf("name_%03d", a)
[1] "name_001" "name_026" "name_051" "name_076" "name_101"
# formatC()
paste("name", formatC(a, width=3, flag="0"), sep="_")
[1] "name_001" "name_026" "name_051" "name_076" "name_101"
sprintf does not print
Use cat() or print() outside sprintf(). sprintf() do not print in a non interactive mode.
cat(sprintf('%5.2f\t%i\n',1.234, l234))
Creating publication quality graphs in R
HDF5 : Hierarchical Data Format
HDF5 is an open binary file format for storing and managing large, complex datasets. The file format was developed by the HDF Group, and is widely used in scientific computing.
- https://en.wikipedia.org/wiki/Hierarchical_Data_Format
- HDF5 tutorial and others
- rhdf5 package
- rhdf5 is used by ARCHS4 where you can download R program that will download hdf5 file storing expression and metadata such as gene ID, sample/GSM ID, tissues, et al.
> h5ls(destination_file) group name otype dclass dim 0 / data H5I_GROUP 1 /data expression H5I_DATASET INTEGER 35238 x 65429 2 / info H5I_GROUP 3 /info author H5I_DATASET STRING 1 4 /info contact H5I_DATASET STRING 1 5 /info creation-date H5I_DATASET STRING 1 6 /info lab H5I_DATASET STRING 1 7 /info version H5I_DATASET STRING 1 8 / meta H5I_GROUP 9 /meta Sample_channel_count H5I_DATASET STRING 65429 10 /meta Sample_characteristics_ch1 H5I_DATASET STRING 65429 11 /meta Sample_contact_address H5I_DATASET STRING 65429 12 /meta Sample_contact_city H5I_DATASET STRING 65429 13 /meta Sample_contact_country H5I_DATASET STRING 65429 14 /meta Sample_contact_department H5I_DATASET STRING 65429 15 /meta Sample_contact_email H5I_DATASET STRING 65429 16 /meta Sample_contact_institute H5I_DATASET STRING 65429 17 /meta Sample_contact_laboratory H5I_DATASET STRING 65429 18 /meta Sample_contact_name H5I_DATASET STRING 65429 19 /meta Sample_contact_phone H5I_DATASET STRING 65429 20 /meta Sample_contact_zip-postal_code H5I_DATASET STRING 65429 21 /meta Sample_data_processing H5I_DATASET STRING 65429 22 /meta Sample_data_row_count H5I_DATASET STRING 65429 23 /meta Sample_description H5I_DATASET STRING 65429 24 /meta Sample_extract_protocol_ch1 H5I_DATASET STRING 65429 25 /meta Sample_geo_accession H5I_DATASET STRING 65429 26 /meta Sample_instrument_model H5I_DATASET STRING 65429 27 /meta Sample_last_update_date H5I_DATASET STRING 65429 28 /meta Sample_library_selection H5I_DATASET STRING 65429 29 /meta Sample_library_source H5I_DATASET STRING 65429 30 /meta Sample_library_strategy H5I_DATASET STRING 65429 31 /meta Sample_molecule_ch1 H5I_DATASET STRING 65429 32 /meta Sample_organism_ch1 H5I_DATASET STRING 65429 33 /meta Sample_platform_id H5I_DATASET STRING 65429 34 /meta Sample_relation H5I_DATASET STRING 65429 35 /meta Sample_series_id H5I_DATASET STRING 65429 36 /meta Sample_source_name_ch1 H5I_DATASET STRING 65429 37 /meta Sample_status H5I_DATASET STRING 65429 38 /meta Sample_submission_date H5I_DATASET STRING 65429 39 /meta Sample_supplementary_file_1 H5I_DATASET STRING 65429 40 /meta Sample_supplementary_file_2 H5I_DATASET STRING 65429 41 /meta Sample_taxid_ch1 H5I_DATASET STRING 65429 42 /meta Sample_title H5I_DATASET STRING 65429 43 /meta Sample_type H5I_DATASET STRING 65429 44 /meta genes H5I_DATASET STRING 35238
Formats for writing/saving and sharing data
Efficiently Saving and Sharing Data in R
Write unix format files on Windows and vice versa
https://stat.ethz.ch/pipermail/r-devel/2012-April/063931.html
with() and within() functions
within() is similar to with() except it is used to create new columns and merge them with the original data sets. See youtube video.
closePr <- with(mariokart, totalPr - shipPr)
head(closePr, 20)
mk <- within(mariokart, {
closePr <- totalPr - shipPr
})
head(mk) # new column closePr
mk <- mariokart
aggregate(. ~ wheels + cond, mk, mean)
# create mean according to each level of (wheels, cond)
aggregate(totalPr ~ wheels + cond, mk, mean)
tapply(mk$totalPr, mk[, c("wheels", "cond")], mean)
stem(): stem-and-leaf plot, bar chart on terminals
- https://en.wikipedia.org/wiki/Stem-and-leaf_display
- https://stackoverflow.com/questions/14736556/ascii-plotting-functions-for-r
- txtplot package
Graphical Parameters, Axes and Text, Combining Plots
15 Questions All R Users Have About Plots
See http://blog.datacamp.com/15-questions-about-r-plots/. This is a tremendous post. It covers the built-in plot() function and ggplot() from ggplot2 package.
- How To Draw An Empty R Plot? plot.new()
- How To Set The Axis Labels And Title Of The R Plots?
- How To Add And Change The Spacing Of The Tick Marks Of Your R Plot? axis()
- How To Create Two Different X- or Y-axes? par(new=TRUE), axis(), mtext()
- How To Add Or Change The R Plot’s Legend? legend()
- How To Draw A Grid In Your R Plot? grid()
- How To Draw A Plot With A PNG As Background? rasterImage() from the png package
- How To Adjust The Size Of Points In An R Plot? cex argument
- How To Fit A Smooth Curve To Your R Data? loess() and lines()
- How To Add Error Bars In An R Plot? arrows()
- How To Save A Plot As An Image On Disc
- How To Plot Two R Plots Next To Each Other? par(mfrow), gridBase package, lattice package
- How To Plot Multiple Lines Or Points? plot(), lines()
- How To Fix The Aspect Ratio For Your R Plots? asp parameter
- What Is The Function Of hjust And vjust In ggplot2?
Scatterplot with the "rug" function
require(stats) # both 'density' and its default method
with(faithful, {
plot(density(eruptions, bw = 0.15))
rug(eruptions)
rug(jitter(eruptions, amount = 0.01), side = 3, col = "light blue")
})
See also the stripchart() function which produces one dimensional scatter plots (or dot plots) of the given data.
Identify/Locate Points in a Scatter Plot
Draw a single plot with two different y-axes
Draw Color Palette
SVG
Embed svg in html
svglite
https://blog.rstudio.org/2016/11/14/svglite-1-2-0/
pdf -> svg
Using Inkscape. See this post.
read.table
clipboard
source("clipboard")
read.table("clipboard")
inline text
mydf <- read.table(header=T, text='
cond yval
A 2
B 2.5
C 1.6
')
http(s) connection
temp = getURL("https://gist.github.com/arraytools/6743826/raw/23c8b0bc4b8f0d1bfe1c2fad985ca2e091aeb916/ip.txt",
ssl.verifypeer = FALSE)
ip <- read.table(textConnection(temp), as.is=TRUE)
read only specific columns
Use 'colClasses' option in read.table, read.delim, .... For example, the following example reads only the 3rd column of the text file and also changes its data type from a data frame to a vector. Note that we have include double quotes around NULL.
x <- read.table("var_annot.vcf", colClasses = c(rep("NULL", 2), "character", rep("NULL", 7)),
skip=62, header=T, stringsAsFactors = FALSE)[, 1]
#
system.time(x <- read.delim("Methylation450k.txt",
colClasses = c("character", "numeric", rep("NULL", 188)), stringsAsFactors = FALSE))
To know the number of columns, we might want to read the first row first.
library(magrittr)
scan("var_annot.vcf", sep="\t", what="character", skip=62, nlines=1, quiet=TRUE) %>% length()
Another method is to use pipe(), cut or awk. See ways to read only selected columns from a file into R
Serialization
If we want to pass an R object to C (use recv() function), we can use writeBin() to output the stream size and then use serialize() function to output the stream to a file. See the post on R mailing list.
> a <- list(1,2,3) > a_serial <- serialize(a, NULL) > a_length <- length(a_serial) > a_length [1] 70 > writeBin(as.integer(a_length), connection, endian="big") > serialize(a, connection)
In C++ process, I receive one int variable first to get the length, and then read <length> bytes from the connection.
socketConnection
See ?socketconnection.
Simple example
from the socketConnection's manual.
Open one R session
con1 <- socketConnection(port = 22131, server = TRUE) # wait until a connection from some client writeLines(LETTERS, con1) close(con1)
Open another R session (client)
con2 <- socketConnection(Sys.info()["nodename"], port = 22131)
# as non-blocking, may need to loop for input
readLines(con2)
while(isIncomplete(con2)) {
Sys.sleep(1)
z <- readLines(con2)
if(length(z)) print(z)
}
close(con2)
Use nc in client
The client does not have to be the R. We can use telnet, nc, etc. See the post here. For example, on the client machine, we can issue
nc localhost 22131 [ENTER]
Then the client will wait and show anything written from the server machine. The connection from nc will be terminated once close(con1) is given.
If I use the command
nc -v -w 2 localhost -z 22130-22135
then the connection will be established for a short time which means the cursor on the server machine will be returned. If we issue the above nc command again on the client machine it will show the connection to the port 22131 is refused. PS. "-w" switch denotes the number of seconds of the timeout for connects and final net reads.
Some post I don't have a chance to read. http://digitheadslabnotebook.blogspot.com/2010/09/how-to-send-http-put-request-from-r.html
Use curl command in client
On the server,
con1 <- socketConnection(port = 8080, server = TRUE)
On the client,
curl --trace-ascii debugdump.txt http://localhost:8080/
Then go to the server,
while(nchar(x <- readLines(con1, 1)) > 0) cat(x, "\n") close(con1) # return cursor in the client machine
Use telnet command in client
On the server,
con1 <- socketConnection(port = 8080, server = TRUE)
On the client,
sudo apt-get install telnet telnet localhost 8080 abcdefg hijklmn qestst
Go to the server,
readLines(con1, 1) readLines(con1, 1) readLines(con1, 1) close(con1) # return cursor in the client machine
Some tutorial about using telnet on http request. And this is a summary of using telnet.
Subsetting
Subset assignment of R Language Definition and Manipulation of functions.
The result of the command x[3:5] <- 13:15 is as if the following had been executed
`*tmp*` <- x x <- "[<-"(`*tmp*`, 3:5, value=13:15) rm(`*tmp*`)
Avoid Coercing Indices To Doubles
as.formula()
? as.formula
xnam <- paste("x", 1:25, sep="")
fmla <- as.formula(paste("y ~ ", paste(xnam, collapse= "+")))
outcome <- "mpg"
variables <- c("cyl", "disp", "hp", "carb")
# Method 1. The 'Call' portion of the model is reported as “formula = f”
# our modeling effort,
# fully parameterized!
f <- as.formula(
paste(outcome,
paste(variables, collapse = " + "),
sep = " ~ "))
print(f)
# mpg ~ cyl + disp + hp + carb
model <- lm(f, data = mtcars)
print(model)
# Call:
# lm(formula = f, data = mtcars)
#
# Coefficients:
# (Intercept) cyl disp hp carb
# 34.021595 -1.048523 -0.026906 0.009349 -0.926863
# Method 2. eval() + bquote() + ".()"
format(terms(model)) # or model$terms
# [1] "mpg ~ cyl + disp + hp + carb"
# The new line of code
model <- eval(bquote( lm(.(f), data = mtcars) ))
print(model)
# Call:
# lm(formula = mpg ~ cyl + disp + hp + carb, data = mtcars)
#
# Coefficients:
# (Intercept) cyl disp hp carb
# 34.021595 -1.048523 -0.026906 0.009349 -0.926863
# Note if we skip ".()" operator
> eval(bquote( lm(f, data = mtcars) ))
Call:
lm(formula = f, data = mtcars)
Coefficients:
(Intercept) cyl disp hp carb
34.021595 -1.048523 -0.026906 0.009349 -0.926863
S3 and S4 methods
- How S4 works in R https://www.rdocumentation.org/packages/methods/versions/3.5.1/topics/Methods_Details
- Software for Data Analysis: Programming with R by John Chambers
- Programming with Data: A Guide to the S Language by John Chambers
- https://www.rmetrics.org/files/Meielisalp2009/Presentations/Chalabi1.pdf
- https://www.stat.auckland.ac.nz/S-Workshop/Gentleman/S4Objects.pdf
- packS4: Toy Example of S4 Package
- http://www.cyclismo.org/tutorial/R/s4Classes.html
- http://adv-r.had.co.nz/S4.html
To get the source code of S4 methods, we can use showMethod(), getMethod() and showMethod(). For example
library(qrqc)
showMethods("gcPlot")
getMethod("gcPlot", "FASTQSummary") # get an error
showMethods("gcPlot", "FASTQSummary") # good.
- getClassDef() in S4 (Bioconductor course).
library(IRanges) ir <- IRanges(start=c(10, 20, 30), width=5) ir class(ir) ## [1] "IRanges" ## attr(,"package") ## [1] "IRanges" getClassDef(class(ir)) ## Class "IRanges" [package "IRanges"] ## ## Slots: ## ## Name: start width NAMES elementType ## Class: integer integer characterORNULL character ## ## Name: elementMetadata metadata ## Class: DataTableORNULL list ## ## Extends: ## Class "Ranges", directly ## Class "IntegerList", by class "Ranges", distance 2 ## Class "RangesORmissing", by class "Ranges", distance 2 ## Class "AtomicList", by class "Ranges", distance 3 ## Class "List", by class "Ranges", distance 4 ## Class "Vector", by class "Ranges", distance 5 ## Class "Annotated", by class "Ranges", distance 6 ## ## Known Subclasses: "NormalIRanges"
See what methods work on an object
see what methods work on an object, e.g. a GRanges object:
methods(class="GRanges")
Or if you have an object, x:
methods(class=class(x))
View S3 function definition: double colon '::' and triple colon ':::' operators
?":::"
- pkg::name returns the value of the exported variable name in namespace pkg
- pkg:::name returns the value of the internal variable name
base::"+" stats:::coef.default
mcols() and DataFrame() from Bioc S4Vectors package
- mcols: Get or set the metadata columns.
- colData: SummarizedExperiment instances from GenomicRanges
- DataFrame: The DataFrame class extends the DataTable virtual class and supports the storage of any type of object (with length and [ methods) as columns.
For example, in Shrinkage of logarithmic fold changes vignette of the DESeq2paper package
> mcols(ddsNoPrior[genes, ])
DataFrame with 2 rows and 21 columns
baseMean baseVar allZero dispGeneEst dispFit dispersion dispIter dispOutlier dispMAP
<numeric> <numeric> <logical> <numeric> <numeric> <numeric> <numeric> <logical> <numeric>
1 163.5750 8904.607 FALSE 0.06263141 0.03862798 0.0577712 7 FALSE 0.0577712
2 175.3883 59643.515 FALSE 2.25306109 0.03807917 2.2530611 12 TRUE 1.6011440
Intercept strain_DBA.2J_vs_C57BL.6J SE_Intercept SE_strain_DBA.2J_vs_C57BL.6J WaldStatistic_Intercept
<numeric> <numeric> <numeric> <numeric> <numeric>
1 6.210188 1.735829 0.1229354 0.1636645 50.515872
2 6.234880 1.823173 0.6870629 0.9481865 9.074686
WaldStatistic_strain_DBA.2J_vs_C57BL.6J WaldPvalue_Intercept WaldPvalue_strain_DBA.2J_vs_C57BL.6J
<numeric> <numeric> <numeric>
1 10.60602 0.000000e+00 2.793908e-26
2 1.92280 1.140054e-19 5.450522e-02
betaConv betaIter deviance maxCooks
<logical> <numeric> <numeric> <numeric>
1 TRUE 3 210.4045 0.2648753
2 TRUE 9 243.7455 0.3248949
findInterval()
Related functions are cuts() and split(). See also
do.call, rbind, lapply
Lots of examples. See for example this one for creating a data frame from a vector.
x <- readLines(textConnection("---CLUSTER 1 ---
3
4
5
6
---CLUSTER 2 ---
9
10
8
11"))
# create a list of where the 'clusters' are
clust <- c(grep("CLUSTER", x), length(x) + 1L)
# get size of each cluster
clustSize <- diff(clust) - 1L
# get cluster number
clustNum <- gsub("[^0-9]+", "", x[grep("CLUSTER", x)])
result <- do.call(rbind, lapply(seq(length(clustNum)), function(.cl){
cbind(Object = x[seq(clust[.cl] + 1L, length = clustSize[.cl])]
, Cluster = .cl
)
}))
result
Object Cluster
[1,] "3" "1"
[2,] "4" "1"
[3,] "5" "1"
[4,] "6" "1"
[5,] "9" "2"
[6,] "10" "2"
[7,] "8" "2"
[8,] "11" "2"
A 2nd example is to sort a data frame by using do.call(order, list()).
How to get examples from help file
See this post. Method 1:
example(acf, give.lines=TRUE)
Method 2:
Rd <- utils:::.getHelpFile(?acf) tools::Rd2ex(Rd)
"[" and "[[" with the sapply() function
Suppose we want to extract string from the id like "ABC-123-XYZ" before the first hyphen.
sapply(strsplit("ABC-123-XYZ", "-"), "[", 1)
is the same as
sapply(strsplit("ABC-123-XYZ", "-"), function(x) x[1])
Dealing with date
d1 = date()
class(d1) # "character"
d2 = Sys.Date()
class(d2) # "Date"
format(d2, "%a %b %d")
library(lubridate); ymd("20140108") # "2014-01-08 UTC"
mdy("08/04/2013") # "2013-08-04 UTC"
dmy("03-04-2013") # "2013-04-03 UTC"
ymd_hms("2011-08-03 10:15:03") # "2011-08-03 10:15:03 UTC"
ymd_hms("2011-08-03 10:15:03", tz="Pacific/Auckland")
# "2011-08-03 10:15:03 NZST"
?Sys.timezone
x = dmy(c("1jan2013", "2jan2013", "31mar2013", "30jul2013"))
wday(x[1]) # 3
wday(x[1], label=TRUE) # Tues
- http://www.r-statistics.com/2012/03/do-more-with-dates-and-times-in-r-with-lubridate-1-1-0/
- http://cran.r-project.org/web/packages/lubridate/vignettes/lubridate.html
- http://rpubs.com/seandavi/GEOMetadbSurvey2014
- We want our dates and times as class "Date" or the class "POSIXct", "POSIXlt". For more information type ?POSIXlt.
Nonstandard evaluation and deparse/substitute
- Vignette from the lazyeval package. It is needed in three cases
- Labelling: turn an argument into a label
- Formulas
- Dot-dot-dot
- substitute(expr, env) - capture expression.
- substitute() is often paired with deparse() to create informative labels for data sets and plots.
- Use 'substitute' to include the variable's name in a plot title, e.g.: var <- "abc"; hist(var,main=substitute(paste("Dist of ", var))) will show the title "Dist of var" instead of "Dist of abc" in the title.
- quote(expr) - similar to substitute() but do nothing??
- eval(expr, envir), evalq(expr, envir) - eval evaluates its first argument in the current scope before passing it to the evaluator: evalq avoids this.
- deparse(expr) - turns unevaluated expressions into character strings. For example,
> deparse(args(lm)) [1] "function (formula, data, subset, weights, na.action, method = \"qr\", " [2] " model = TRUE, x = FALSE, y = FALSE, qr = TRUE, singular.ok = TRUE, " [3] " contrasts = NULL, offset, ...) " [4] "NULL" > deparse(args(lm), width=20) [1] "function (formula, data, " " subset, weights, " [3] " na.action, method = \"qr\", " " model = TRUE, x = FALSE, " [5] " y = FALSE, qr = TRUE, " " singular.ok = TRUE, " [7] " contrasts = NULL, " " offset, ...) " [9] "NULL"
- parse(text) - returns the parsed but unevaluated expressions in a list. See Create a Simple Socket Server in R for the application of eval(parse(text)). Be cautious!
Following is another example. Assume we have a bunch of functions (f1, f2, ...; each function implements a different algorithm) with same input arguments format (eg a1, a2). We like to run these function on the same data (to compare their performance).
f1 <- function(x) x+1; f2 <- function(x) x+2; f3 <- function(x) x+3
f1(1:3)
f2(1:3)
f3(1:3)
# Or
myfun <- function(f, a) {
eval(parse(text = f))(a)
}
myfun("f1", 1:3)
myfun("f2", 1:3)
myfun("f3", 1:3)
# Or with lapply
method <- c("f1", "f2", "f3")
res <- lapply(method, function(M) {
Mres <- eval(parse(text = M))(1:3)
return(Mres)
})
names(res) <- method
The ‘…’ argument
See Section 10.4 of An Introduction to R. Especially, the expression list(...) evaluates all such arguments and returns them in a named list
Lazy evaluation in R functions arguments
- http://adv-r.had.co.nz/Functions.html
- https://stat.ethz.ch/pipermail/r-devel/2015-February/070688.html
R function arguments are lazy — they’re only evaluated if they’re actually used.
- Example 1. By default, R function arguments are lazy.
f <- function(x) {
999
}
f(stop("This is an error!"))
#> [1] 999
- Example 2. If you want to ensure that an argument is evaluated you can use force().
add <- function(x) {
force(x)
function(y) x + y
}
adders2 <- lapply(1:10, add)
adders2[[1]](10)
#> [1] 11
adders2[[10]](10)
#> [1] 20
- Example 3. Default arguments are evaluated inside the function.
f <- function(x = ls()) {
a <- 1
x
}
# ls() evaluated inside f:
f()
# [1] "a" "x"
# ls() evaluated in global environment:
f(ls())
# [1] "add" "adders" "f"
- Example 4. Laziness is useful in if statements — the second statement below will be evaluated only if the first is true.
x <- NULL
if (!is.null(x) && x > 0) {
}
Backtick sign, infix/prefix/postfix operators
The backtick sign ` (not the single quote) refers to functions or variables that have otherwise reserved or illegal names; e.g. '&&', '+', '(', 'for', 'if', etc. See some examples in this note.
infix operator.
1 + 2 # infix + 1 2 # prefix 1 2 + # postfix
List data type
Calling a function given a list of arguments
> args <- list(c(1:10, NA, NA), na.rm = TRUE) > do.call(mean, args) [1] 5.5 > mean(c(1:10, NA, NA), na.rm = TRUE) [1] 5.5
Error handling and exceptions, tryCatch(), stop(), warning() and message()
- http://adv-r.had.co.nz/Exceptions-Debugging.html
- try() allows execution to continue even after an error has occurred. You can suppress the message with try(..., silent = TRUE).
out <- try({
a <- 1
b <- "x"
a + b
})
elements <- list(1:10, c(-1, 10), c(T, F), letters)
results <- lapply(elements, log)
is.error <- function(x) inherits(x, "try-error")
succeeded <- !sapply(results, is.error)
- tryCatch(): With tryCatch() you map conditions to handlers (like switch()), named functions that are called with the condition as an input. Note that try() is a simplified version of tryCatch().
tryCatch(expr, ..., finally)
show_condition <- function(code) {
tryCatch(code,
error = function(c) "error",
warning = function(c) "warning",
message = function(c) "message"
)
}
show_condition(stop("!"))
#> [1] "error"
show_condition(warning("?!"))
#> [1] "warning"
show_condition(message("?"))
#> [1] "message"
show_condition(10)
#> [1] 10
Below is another snippet from available.packages() function,
z <- tryCatch(download.file(....), error = identity) if (!inherits(z, "error")) STATEMENTS
Using list type
Avoid if-else or switch
?plot.stepfun.
y0 <- c(1,2,4,3)
sfun0 <- stepfun(1:3, y0, f = 0)
sfun.2 <- stepfun(1:3, y0, f = .2)
sfun1 <- stepfun(1:3, y0, right = TRUE)
tt <- seq(0, 3, by = 0.1)
op <- par(mfrow = c(2,2))
plot(sfun0); plot(sfun0, xval = tt, add = TRUE, col.hor = "bisque")
plot(sfun.2);plot(sfun.2, xval = tt, add = TRUE, col = "orange") # all colors
plot(sfun1);lines(sfun1, xval = tt, col.hor = "coral")
##-- This is revealing :
plot(sfun0, verticals = FALSE,
main = "stepfun(x, y0, f=f) for f = 0, .2, 1")
for(i in 1:3)
lines(list(sfun0, sfun.2, stepfun(1:3, y0, f = 1))[[i]], col = i)
legend(2.5, 1.9, paste("f =", c(0, 0.2, 1)), col = 1:3, lty = 1, y.intersp = 1)
par(op)
Open a new Window device
X11() or dev.new()
par()
?par
text size and font on main, lab & axis
- Graphical Parameters from statmethods.net.
- Overlaying Data Summaries in Dotplots
Examples:
- cex.main=0.9
- cex.lab=0.8
- font.lab=2
- cex.axis=0.8
- font.axis=2
- col.axis="grey50"
layout
http://datascienceplus.com/adding-text-to-r-plot/
reset the settings
op <- par(mfrow=c(2,1), mar = c(5,7,4,2) + 0.1) .... par(op) # mfrow=c(1,1), mar = c(5,4,4,2) + .1
mtext (margin text) vs title
- https://datascienceplus.com/adding-text-to-r-plot/
- https://datascienceplus.com/mastering-r-plot-part-2-axis/
mgp (axis label locations)
- The margin line (in ‘mex’ units) for the axis title, axis labels and axis line. Note that ‘mgp[1]’ affects ‘title’ whereas ‘mgp[2:3]’ affect ‘axis’. The default is ‘c(3, 1, 0)’. If we like to make the axis labels closer to an axis, we can use mgp=c(2.3, 1, 0) for example.
- http://rfunction.com/archives/1302 mgp – A numeric vector of length 3, which sets the axis label locations relative to the edge of the inner plot window. The first value represents the location the labels (i.e. xlab and ylab in plot), the second the tick-mark labels, and third the tick marks. The default is c(3, 1, 0).
pch
- Full circle: pch=16
lty (line type)
las (label style)
0: The default, parallel to the axis
1: Always horizontal
2: Perpendicular to the axis
3: Always vertical
oma (outer margin), common title for two plots
The following trick is useful when we want to draw multiple plots with a common title.
par(mfrow=c(1,2),oma = c(0, 0, 2, 0)) # oma=c(0, 0, 0, 0) by default
plot(1:10, main="Plot 1")
plot(1:100, main="Plot 2")
mtext("Title for Two Plots", outer = TRUE, cex = 1.5) # outer=FALSE by default
Mastering R plot – Part 3: Outer margins mtext() & par(xpd).
Non-standard fonts in postscript and pdf graphics
https://cran.r-project.org/doc/Rnews/Rnews_2006-2.pdf#page=41
Suppress warnings
Use options(). If warn is negative all warnings are ignored. If warn is zero (the default) warnings are stored until the top--level function returns.
op <- options("warn")
options(warn = -1)
....
options(op)
# OR
warnLevel <- options()$warn
options(warn = -1)
...
options(warn = warnLevel)
NULL, NA, NaN, Inf
https://tomaztsql.wordpress.com/2018/07/04/r-null-values-null-na-nan-inf/
save() vs saveRDS()
- saveRDS() can only save one R object while save() does not have this constraint.
- saveRDS() doesn’t save the both the object and its name it just saves a representation of the object. As a result, the saved object can be loaded into a named object within R that is different from the name it had when originally serialized. See this post.
x <- 5
saveRDS(x, "myfile.rds")
x2 <- readRDS("myfile.rds")
identical(mod, mod2, ignore.environment = TRUE)
==, all.equal(), identical()
- ==: exact match
- all.equal: compare R objects x and y testing ‘near equality’
- identical: The safe and reliable way to test two objects for being exactly equal.
x <- 1.0; y <- 0.99999999999 all.equal(x, y) # [1] TRUE identical(x, y) # [1] FALSE
See also the testhat package.
Numerical Pitfall
Numerical pitfalls in computing variance
.1 - .3/3 ## [1] 0.00000000000000001388
Sys.getpid()
This can be used to monitor R process memory usage or stop the R process. See this post.
How to debug an R code
Using assign() in functions
For example, insert the following line to your function
assign(envir=globalenv(), "GlobalVar", localvar)
Debug lapply()/sapply()
- https://stackoverflow.com/questions/1395622/debugging-lapply-sapply-calls
- https://stat.ethz.ch/R-manual/R-devel/library/utils/html/recover.html. Use options(error=NULL) to turn it off.
Debugging with RStudio
- https://www.rstudio.com/resources/videos/debugging-techniques-in-rstudio/
- https://github.com/ajmcoqui/debuggingRStudio/blob/master/RStudio_Debugging_Cheatsheet.pdf
- https://support.rstudio.com/hc/en-us/articles/205612627-Debugging-with-RStudio
Debug R source code
Build R with debug information
- R -> Build R from its source on Windows
- http://www.stats.uwo.ca/faculty/murdoch/software/debuggingR/
- http://www.stats.uwo.ca/faculty/murdoch/software/debuggingR/gdb.shtml
- My note of debugging cor() function
.Call
- Writing R Extensions manual.
Registering native routines
https://cran.rstudio.com/doc/manuals/r-release/R-exts.html#Registering-native-routines
Pay attention to the prefix argument .fixes (eg .fixes = "C_") in useDynLib() function in the NAMESPACE file.
Example of debugging cor() function
Note that R's cor() function called a C function cor().
stats::cor .... .Call(C_cor, x, y, na.method, method == "kendall")
A step-by-step screenshot of debugging using the GNU debugger gdb can be found on my Github repository https://github.com/arraytools/r-debug.
Locale bug (grep did not handle UTF-8 properly PR#16264)
https://bugs.r-project.org/bugzilla3/show_bug.cgi?id=16264
Path length in dir.create() (PR#17206)
https://bugs.r-project.org/bugzilla3/show_bug.cgi?id=17206 (Windows only)
install.package() error, R_LIBS_USER is empty in R 3.4.1
- https://support.rstudio.com/hc/en-us/community/posts/115008369408-Since-update-to-R-3-4-1-R-LIBS-USER-is-empty and http://r.789695.n4.nabble.com/R-LIBS-USER-on-Ubuntu-16-04-td4740935.html. Modify /etc/R/Renviron (if you have a sudo right) by uncomment out line 43.
R_LIBS_USER=${R_LIBS_USER-'~/R/x86_64-pc-linux-gnu-library/3.4'}
- https://stackoverflow.com/questions/44873972/default-r-personal-library-location-is-null. Modify $HOME/.Renviron by adding a line
R_LIBS_USER="${HOME}/R/${R_PLATFORM}-library/3.4"
- http://stat.ethz.ch/R-manual/R-devel/library/base/html/libPaths.html. Play with .libPaths()
On Mac & R 3.4.0 (it's fine)
> Sys.getenv("R_LIBS_USER")
[1] "~/Library/R/3.4/library"
> .libPaths()
[1] "/Library/Frameworks/R.framework/Versions/3.4/Resources/library"
On Linux & R 3.3.1 (ARM)
> Sys.getenv("R_LIBS_USER")
[1] "~/R/armv7l-unknown-linux-gnueabihf-library/3.3"
> .libPaths()
[1] "/home/$USER/R/armv7l-unknown-linux-gnueabihf-library/3.3"
[2] "/usr/local/lib/R/library"
On Linux & R 3.4.1 (*Problem*)
> Sys.getenv("R_LIBS_USER")
[1] ""
> .libPaths()
[1] "/usr/local/lib/R/site-library" "/usr/lib/R/site-library"
[3] "/usr/lib/R/library"
I need to specify the lib parameter when I use the install.packages command.
> install.packages("devtools", "~/R/x86_64-pc-linux-gnu-library/3.4")
> library(devtools)
Error in library(devtools) : there is no package called 'devtools'
# Specify lib.loc parameter will not help with the dependency package
> library(devtools, lib.loc = "~/R/x86_64-pc-linux-gnu-library/3.4")
Error: package or namespace load failed for 'devtools':
.onLoad failed in loadNamespace() for 'devtools', details:
call: loadNamespace(name)
error: there is no package called 'withr'
# A solution is to redefine .libPaths
> .libPaths(c("~/R/x86_64-pc-linux-gnu-library/3.4", .libPaths()))
> library(devtools) # Works
A better solution is to specify R_LIBS_USER in ~/.Renviron file or ~/.bash_profile; see ?Startup.
Using external data from within another package
https://logfc.wordpress.com/2017/03/02/using-external-data-from-within-another-package/
How to run R scripts from the command line
https://support.rstudio.com/hc/en-us/articles/218012917-How-to-run-R-scripts-from-the-command-line
How to exit a sourced R script
- How to exit a sourced R script
- Problem using the source-function within R-functions The best way to handle the generic sort of problem you are describing is to take those source'd files, and rewrite their content as functions to be called from your other functions.
Decimal point & decimal comma
Countries using Arabic numerals with decimal comma (Austria, Belgium, Brazil France, Germany, Netherlands, Norway, South Africa, Spain, Sweden, ...) https://en.wikipedia.org/wiki/Decimal_mark
setting seed locally (not globally) in R
https://stackoverflow.com/questions/14324096/setting-seed-locally-not-globally-in-r
R's internal C API
https://github.com/hadley/r-internals
Random numbers: multivariate normal
Why MASS::mvrnorm() gives different result on Mac and Linux/Windows?
The reason could be the covariance matrix decomposition - and that may be due to the LAPACK/BLAS libraries. See
- https://stackoverflow.com/questions/11567613/different-random-number-generation-between-os
- https://stats.stackexchange.com/questions/149321/generating-and-working-with-random-vectors-in-r
- Cholesky versus eigendecomposition for drawing samples from a multivariate normal distribution
set.seed(1234)
junk <- biospear::simdata(n=500, p=500, q.main = 10, q.inter = 10,
prob.tt = .5, m0=1, alpha.tt= -.5,
beta.main= -.5, beta.inter= -.5, b.corr = .7, b.corr.by=25,
wei.shape = 1, recr=3, fu=2, timefactor=1)
## Method 1: MASS::mvrnorm()
## This is simdata() has used. It gives different numbers on different OS.
##
library(MASS)
set.seed(1234)
m0 <-1
n <- 500
prob.tt <- .5
p <- 500
b.corr.by <- 25
b.corr <- .7
data <- data.frame(treat = rbinom(n, 1, prob.tt) - 0.5)
n.blocks <- p%/%b.corr.by
covMat <- diag(n.blocks) %x%
matrix(b.corr^abs(matrix(1:b.corr.by, b.corr.by, b.corr.by, byrow = TRUE) -
matrix(1:b.corr.by, b.corr.by, b.corr.by)), b.corr.by, b.corr.by)
diag(covMat) <- 1
data <- cbind(data, mvrnorm(n, rep(0, p), Sigma = covMat))
range(data)
# Mac: -4.963827 4.133723
# Linux/Windows: -4.327635 4.408097
packageVersion("MASS")
# Mac: [1] ‘7.3.49’
# Linux: [1] ‘7.3.49’
# Windows: [1] ‘7.3.47’
R.version$version.string
# Mac: [1] "R version 3.4.3 (2017-11-30)"
# Linux: [1] "R version 3.4.4 (2018-03-15)"
# Windows: [1] "R version 3.4.3 (2017-11-30)"
## Method 2: mvtnorm::rmvnorm()
library(mvtnorm)
set.seed(1234)
sigma <- matrix(c(4,2,2,3), ncol=2)
x <- rmvnorm(n=n, rep(0, p), sigma=covMat)
range(x)
# Mac: [1] -4.482566 4.459236
# Linux: [1] -4.482566 4.459236
## Method 3: mvnfast::rmvn()
set.seed(1234)
x <- mvnfast::rmvn(n, rep(0, p), covMat)
range(x)
# Mac: [1] -4.323585 4.355666
# Linux: [1] -4.323585 4.355666
library(microbenchmark)
library(MASS)
library(mvtnorm)
library(mvnfast)
microbenchmark(v1 <- rmvnorm(n=n, rep(0, p), sigma=covMat, "eigen"),
v2 <- rmvnorm(n=n, rep(0, p), sigma=covMat, "svd"),
v3 <- rmvnorm(n=n, rep(0, p), sigma=covMat, "chol"),
v4 <- rmvn(n, rep(0, p), covMat),
v5 <- mvrnorm(n, rep(0, p), Sigma = covMat))
Unit: milliseconds
expr min lq
v1 <- rmvnorm(n = n, rep(0, p), sigma = covMat, "eigen") 296.55374 300.81089
v2 <- rmvnorm(n = n, rep(0, p), sigma = covMat, "svd") 461.81867 466.98806
v3 <- rmvnorm(n = n, rep(0, p), sigma = covMat, "chol") 118.33759 120.01829
v4 <- rmvn(n, rep(0, p), covMat) 66.64675 69.89383
v5 <- mvrnorm(n, rep(0, p), Sigma = covMat) 291.19826 294.88038
mean median uq max neval cld
306.72485 301.99339 304.46662 335.6137 100 d
478.58536 470.44085 493.89041 571.7990 100 e
125.85427 121.26185 122.21361 151.1658 100 b
71.67996 70.52985 70.92923 100.2622 100 a
301.88144 296.76028 299.50839 346.7049 100 c
A little more investigation shows the eigen values differ a little bit on macOS and Linux.
set.seed(1234); x <- mvrnorm(n, rep(0, p), Sigma = covMat) debug(mvrnorm) # eS --- macOS # eS2 -- Linux Browse[2]> range(abs(eS$values - eS2$values)) # [1] 0.000000e+00 1.776357e-15 Browse[2]> var(as.vector(eS$vectors)) [1] 0.002000006 Browse[2]> var(as.vector(eS2$vectors)) [1] 0.001999987 Browse[2]> all.equal(eS$values, eS2$values) [1] TRUE Browse[2]> which(eS$values != eS2$values) [1] 6 7 8 9 10 11 12 13 14 20 22 23 24 25 26 27 28 29 ... [451] 494 495 496 497 499 500 Browse[2]> range(abs(eS$vectors - eS2$vectors)) [1] 0.0000000 0.5636919
rle() running length encoding
- https://en.wikipedia.org/wiki/Run-length_encoding
- How to Find Consecutive Repeats in R
- R Function of the Day: rle
- Creating nice tables using R Markdown
- https://rosettacode.org/wiki/Run-length_encoding
- R's base::rle() function
- R's Rle class from S4Vectors package which was used in for example IRanges/GRanges/GenomicRanges package
Resource
Books
- A list of recommended books http://blog.revolutionanalytics.com/2015/11/r-recommended-reading.html
- Learning R programming by reading books: A book list
- Modern Applied Statistics with S by William N. Venables and Brian D. Ripley
- Seamless R and C++ Integration with Rcpp by Dirk Eddelbuettel
- Advanced R by Hadley Wickham 2014
- http://brettklamer.com/diversions/statistical/compile-hadleys-advanced-r-programming-to-a-pdf/ Compile Hadley's Advanced R to a PDF
- Functional programming and unit testing for data munging with R by Bruno Rodrigues
- R Cookbook by Paul Teetor
- Machine Learning with R by Brett Lantz
- R for Everyone by Jared P. Lander
- The Art of R Programming by Norman Matloff
- Applied Predictive Modeling by Max Kuhn
- R in Action by Robert Kabacoff
- The R Book by Michael J. Crawley
- Regression Modeling Strategies, with Applications to Linear Models, Survival Analysis and Logistic Regression by Frank E. Harrell
- Data Manipulation with R by Phil Spector
- Review of Efficient R Programming
- R packages: Organize, Test, Document, and Share Your Code by Hadley Wicklam 2015
- Text Mining with R: A Tidy Approach and a blog
- Efficient R programming by Colin Gillespie and Robin Lovelace. It works to re-create the html version of the book if we follow their simple instruction in the Appendix. Note that pdf version has advantages of expected output (mathematical notations, tables) over the epub version.
# R 3.4.1
.libPaths(c("~/R/x86_64-pc-linux-gnu-library/3.4", .libPaths()))
setwd("/tmp/efficientR/")
bookdown::render_book("index.Rmd", output_format = "bookdown::pdf_book")
# generated pdf file is located _book/_main.pdf
bookdown::render_book("index.Rmd", output_format = "bookdown::epub_book")
# generated epub file is located _book/_main.epub.
# This cannot be done in RStudio ("parse_dt" not resolved from current namespace (lubridate))
# but it is OK to run in an R terminal
Webinar
useR!
Blogs, Tips, Socials, Communities
- Google: revolutionanalytics In case you missed it
- Why R is hard to learn by Bob Musenchen.
- My 15 practical tips for a bioinformatician
- The R community is one of R's best features
- Bioinformatics Training at the Harvard Chan Bioinformatics Core
Bug Tracking System
https://bugs.r-project.org/bugzilla3/ and Search existing bug reports. Remember to select 'All' in the Status drop-down list.
Use sessionInfo().